Future Climate Alters Pathogens-Microbiome Co-occurrence Networks in Wheat Straw Residues during Decomposition †
Abstract
:1. Introduction
2. Methods
2.1. Study Site, Experimental Setup, and Design
2.2. Litterbag Preparation, Field Incorporation, and Sampling
2.3. DNA Extraction, PCR, and Illumina Miseq Sequencing
2.4. Sequence Processing and Statistics
3. Results
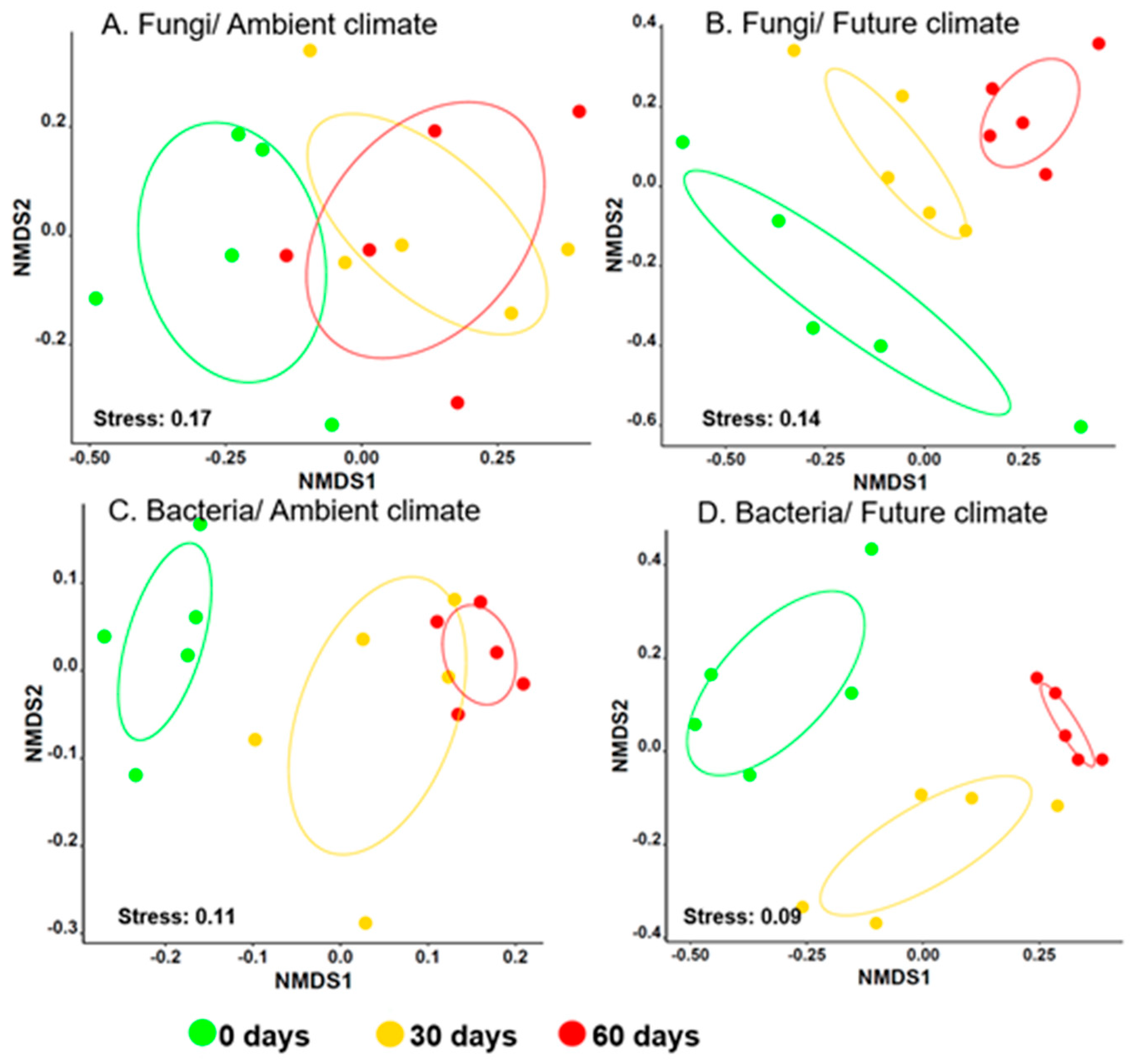
3.1. Climate Change Significantly Alters Microbial Community Composition during Early Stages of Wheat Straw Decomposition
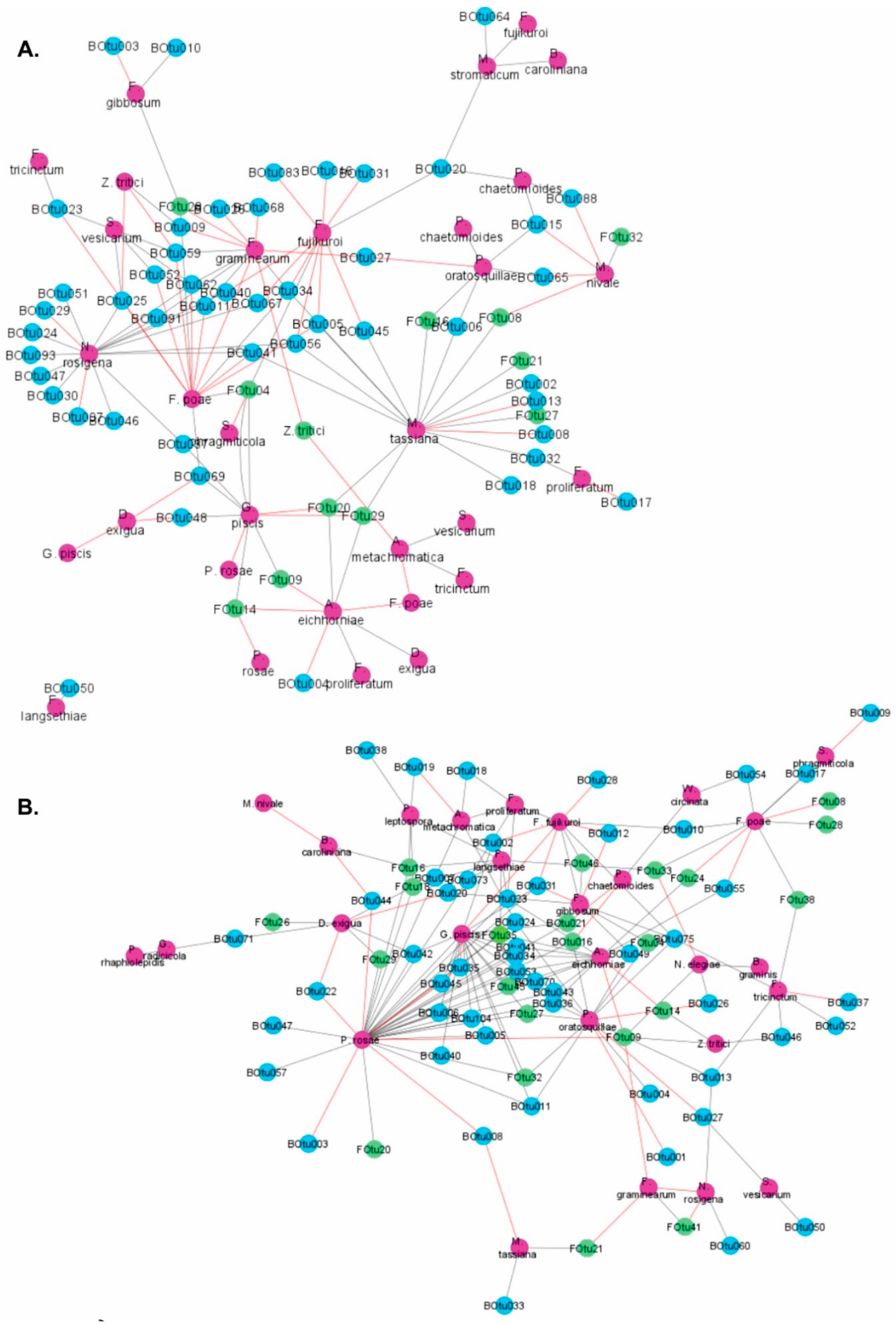
3.2. Co-Occurrence Patterns between Microbes Colonizing Wheat Straw Residues
4. Conclusions
Acknowledgments
References
- Krishna, M.P.; Mohan, M. Litter decomposition in forest ecosystems: A review. Energy Ecol. Environ. 2017, 2, 236–249. [Google Scholar] [CrossRef]
- Kerdraon, L.; Laval, V.; Suffert, F. Microbiomes and pathogen survival in crop residues, an ecotone between plant and soil. Phytobiomes J. 2019, 3, 246–255. [Google Scholar] [CrossRef]
- Duba, A.; Goriewa-Duba, K.; Wachowska, U. A Review of the Interactions between Wheat and Wheat Pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum. Int. J. Mol. Sci. 2018, 19, 1138. [Google Scholar] [CrossRef] [PubMed]
- Kerdraon, L.; Barret, M.; Laval, V.; Suffert, F. Differential dynamics of microbial community networks help identify microorganisms interacting with residue-borne pathogens: The case of Zymoseptoria tritici in wheat. Microbiome 2019, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Fareed Mohamed Wahdan, S.; Hossen, S.; Tanunchai, B.; Schädler, M.; Buscot, F.; Purahong, W. Future climate significantly alters fungal plant pathogen dynamics during the early phase of wheat litter decomposition. Microorganisms 2020, 8, 908. [Google Scholar] [CrossRef] [PubMed]
- Schädler, M.; Buscot, F.; Klotz, S.; Reitz, T.; Durka, W.; Bumberger, J.; Merbach, I.; Michalski, S.G.; Kirsch, K.; Remmler, P.; et al. Investigating the consequences of climate change under different land-use regimes: A novel experimental infrastructure. Ecosphere 2019, 10, e02635. [Google Scholar] [CrossRef]
- Chelius, M.K.; Triplett, E.W. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Bodenhausen, N.; Horton, M.W.; Bergelson, J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 2013, 8, e56329. [Google Scholar] [CrossRef] [PubMed]
- Ihrmark, K.; Bodeker, I.T.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandstrom-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol 2001, 26, 32–46. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wahdan, S.F.M.; Buscot, F.; Purahong, W. Future Climate Alters Pathogens-Microbiome Co-occurrence Networks in Wheat Straw Residues during Decomposition. Proceedings 2020, 66, 22. https://doi.org/10.3390/proceedings2020066022
Wahdan SFM, Buscot F, Purahong W. Future Climate Alters Pathogens-Microbiome Co-occurrence Networks in Wheat Straw Residues during Decomposition. Proceedings. 2020; 66(1):22. https://doi.org/10.3390/proceedings2020066022
Chicago/Turabian StyleWahdan, Sara Fareed Mohamed, François Buscot, and Witoon Purahong. 2020. "Future Climate Alters Pathogens-Microbiome Co-occurrence Networks in Wheat Straw Residues during Decomposition" Proceedings 66, no. 1: 22. https://doi.org/10.3390/proceedings2020066022
APA StyleWahdan, S. F. M., Buscot, F., & Purahong, W. (2020). Future Climate Alters Pathogens-Microbiome Co-occurrence Networks in Wheat Straw Residues during Decomposition. Proceedings, 66(1), 22. https://doi.org/10.3390/proceedings2020066022





