1. Introduction
Pathogenic bacterial contamination in food is a public health concern. It represents a health and safety consumer risk that could cause several diseases and even death. Detection of bacteria is crucial not only in food quality assessment, but for clinical diagnostics and environmental monitoring [
1,
2,
3,
4,
5,
6,
7]. The most commonly used methods for pathogenic detection are polymerase chain reaction (PCR), enzyme linked immunosorbent assay (ELISA) and culture-based assays on differential culture media. Although these methods provide very accurate results, they require specialized technical skills, high-cost, cumbersome procedures time consuming labor (in case of culture-based assay, up to 72 h) [
1,
2,
5,
8,
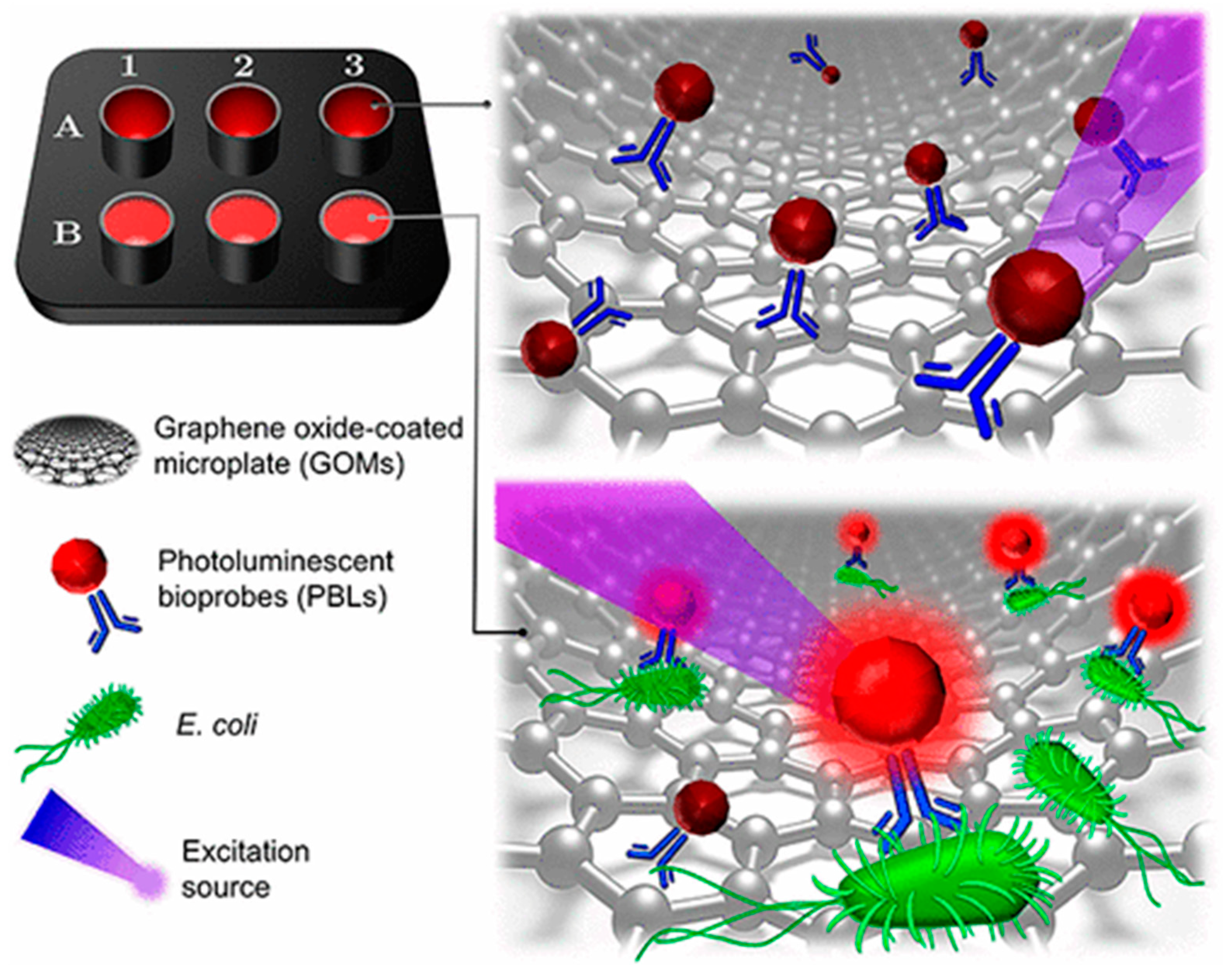
9]. To offer a solution against the disadvantages of these methods, some biosensing systems based on nanomaterials had been developed. We proposed a single-step bacterial detection platform based on graphene oxide coated microplates (GOMs). Non-radiative energy transfer between GOMs and photoluminescence bioprobes (PLBs) is presented in absence of the target analyte, but in presence of analyte, PLBs exhibit strong photoluminescence due to the distance between GOMs and PLBs. These PLBs are a quantum dot-antibody complex, thereby resulting as a biorecognition and interrogation element (see
Figure 1). We explained this behavior between GOMs and PLBs because of the distance between the complex (PLB-bacteria) and GOMs, and because the low affinity between the complex and the GO. Furthermore, industrial food samples were analyzed, such as cauliflower, from a frozen vegetable processor company.
2. Materials and Methods
Sterilized culture-treated 96 well microplates were purchased from Corning, monolayer GO aqueous suspension was purchased from Angstron Materials, PBS tablets and Tween 20 from Sigma-Aldrich, bovine serum albumin (BSA) from Sigma-Aldrich, biotinylate polyclonal Anti-E. coli was purchased from Abcam, Qdot™ 655 Streptavidin Conjugate were purchased from Invitrogen. GO dilutions and buffers were prepared with fresh ultrapure water, Milli-Q system from Millipore. Immunobuffer was prepared using PBS supplemented with 0.5% Tween 20 (v/v) and 1% of BSA (w/v). Standard samples of bacteria were diluted in PBS. The photoluminescence experiments were performed using a Cytation 5 multimode reader (BioTek, Winooski, VT, USA).
GOMs preparation: Microwell plates are easily decorated with GO via hydrophilic interactions, since GO has hydrophilic, so does the microplates. A total of 100 µL of the suspension, with an appropriated concentration, are incubated overnight in the microplate, then, to remove those GO submicrometric sheets that were not attached onto the microplates, washed at least 3 times with ultrapure water.
PLBs preparation: This conjugation was performed by mixing 100 μL of streptavidin-QDs with 100 μL of biotinylated anti-E. coli. These reagents were diluted in immunobuffer, and the reaction was performed during 45 min of mixing at 650 rpm.
Single-step test: Once the microwell plate coated with GO and Ab-QD is prepared the immunoassay requires a single step, deposit 100 µL of sample and 100 µL of Ab-QD in a microwell. The resulting photoluminescence was then recorded throughout 120 min (excitation wavelength: 365 nm, emission wavelength 660 ± 20 nm). Typically, at least three parallel experiments in the same conditions were performed to evaluate precision. The limit of detection was calculated as the average of the control sample added to three times the standard deviation.
Preparation of industrial sample: Industrial samples of cauliflower were kindly provided by La Proxima Estación S.P.R. de R.L. Prior to the analysis the samples were diluted in PBS. Furthermore, these samples were analyzed using 3 M Petrifilm E. coli/Coliform Count (EC) Plates, before being analyzed with the proposed technology.
3. Results
Each biomaterial (antibody) and nanomaterial (GO and QDs) were optimized in terms of concentration to reach an adequate performance of the biosensing system. Then, the platform was characterized. Once the bacterial detection platform was optimized and characterized, we investigated the specificity of the platform. Furthermore, finally, we test the analytical behavior with some industrial food samples.
3.1. Optimización of the Reagents and Caracterization
We decorated GOMs with different concentrations of GO (from 1100 to 1600 μg mL
−1) to find the concentration maximizing modulation of the photoluminescent quenching ratio If/I0; where If is the photoluminescence intensity of PLBs or bare QDs at time x (generally from 5 to 120 min), and I0 is the photoluminescence intensity of PLBs at time 0. This range of concentrations was previously reported by our research group [
10]. We found that the best behavior was with [GO] = 1200 μg mL
−1.
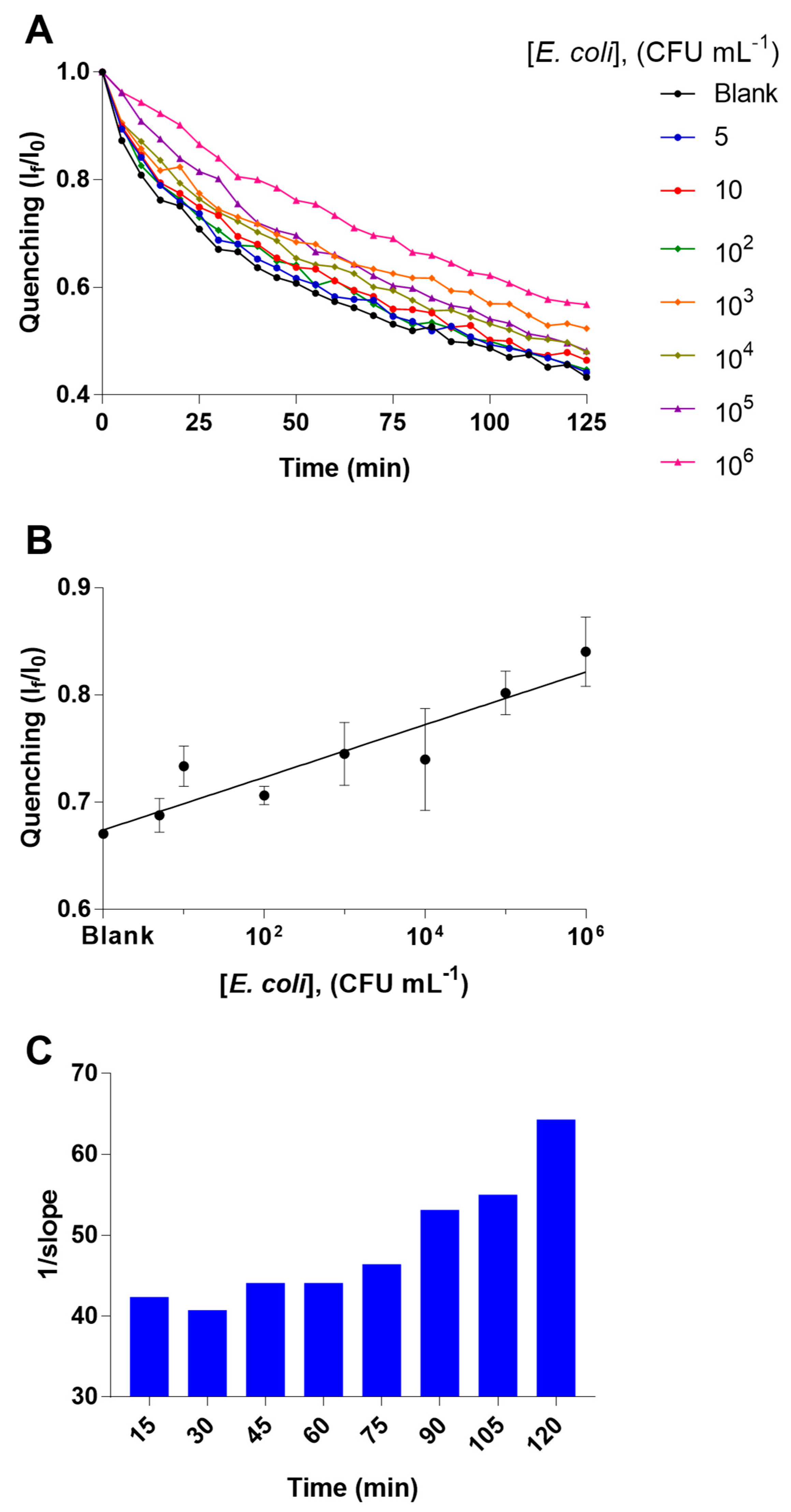
To optimize the analytical behavior of the studied bacterial detection platform in terms sensitivity, we explored the performance of the biosensing system using different concentrations of the PLBs. Particularly, PLBs with a final concentration of QDs-antibody at 0.1125 nM and 0.9 μg mL
−1, respectively, were observed to offer the best analytical performance in terms of both sensitivity and coefficient of variation for the analysis of samples of
E. coli with concentrations from 5 to 10
6 CFU mL
−1. This behavior can be observed in
Figure 2.
3.2. Study of Specificity
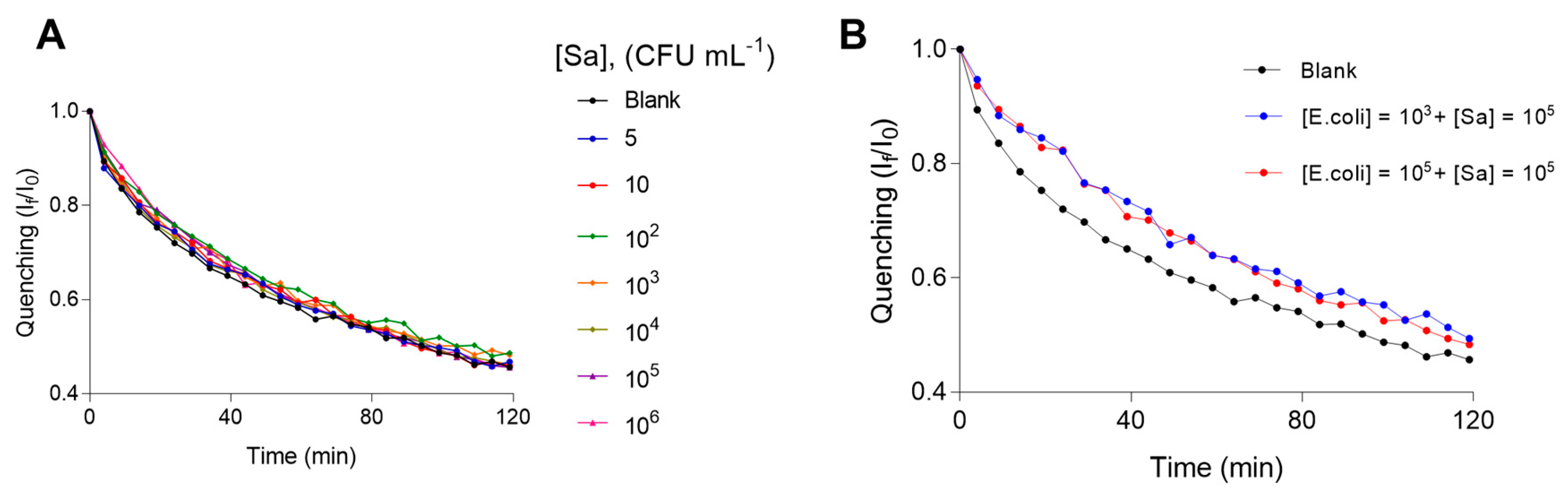
Additionally, we investigated the specificity of the proposed bacterial detection platform by using Salmonella typhimurium. We analyzed different concentrations of the interference bacteria (from 5 to 10
6 CFU mL
−1). We also analyzed other samples containing a relatively low concentration of
E. coli (10
3 CFU mL
−1) and a relatively high concentration of Salmonella (10
5 CFU mL
−1), other with a relatively high concentrations of both strains (10
5 CFU mL
−1). The results of these tests can be observed in
Figure 3A,B.
3.3. Industrial Food Samples
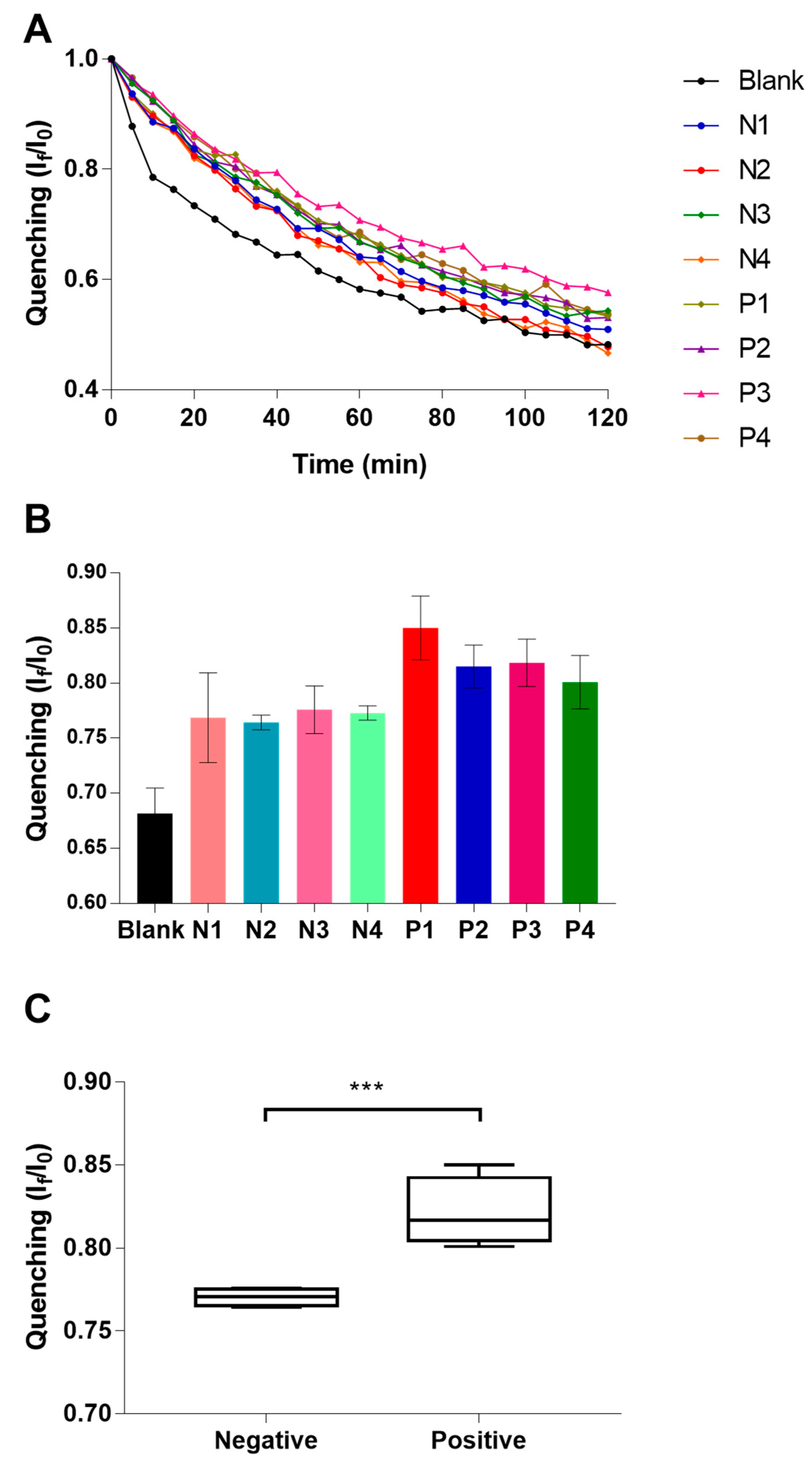
In order to test the feasibility of the proposed technology in the real world, we analyzed industrial samples of cauliflower, these samples were previously determined as positives or negatives via a culture-based method. Particularly, we explored the performance of the real-time quenching profile of four positive samples and four negative samples, respectively (see
Figure 4A–C).
4. Discussion
According to the behavior of the platform in terms of quenching profile (If/I0), we determined that microplate coated with GO 1200 µg mL
−1 shows the best performance for the real-time interrogation of the bacterial detection. Then, different concentrations of PLBs were studied in GOMs prepared with GO 1200 µg mL
−1 and we include some blank sample analysis as control. PLBs with a final concentration of QDs-antibody at 0.1125 nM and 0.9 μg mL
−1, respectively, were observed to offer the best analytical performance in terms of both sensitivity and coefficient of variation. Although there are some concentrations (10 and 10
2 CFU mL
−1) that do not behave as we expected, the precision limits are acceptable. In optimal conditions, the bacterial detection platform reach a limit of detection of 2 CFU mL
−1 at 30 min, reducing the time analysis regarding culture-based methods and improving sensitivity regarding ELISA (review
Figure 2C). For specificity assay, the quenching profile of samples of Salmonella are very similar to blank sample, which confirm the selective character of this novel bacterial detection platform. In the industrial food sample analysis, we could observe that despite the inner matrix effect of the sample, the proposed technology is able to discriminate the negative samples from the positives. Then, analyzed statistically the obtained If/I0 values the two groups of samples. These two groups of samples result statistically different, reaching the lower P value at 30 min of the assay (P = 0.004), it is shown in
Figure 4C.
5. Conclusions
We developed a novel optical platform based on GOMs to pathogenic bacterial determination; this technology operates with a single antibody, avoids wash stages and cumbersome procedures, is not time consuming, and is highly sensitive (around 2 CFU mL−1). For industrial samples, the bacterial detection platform is capable of classifying negative and positive samples in around 30 min. This biosensing strategy potentially offers a cheaper and faster option for the food industry to assure the quality of products and consumer safety.
Author Contributions
(M.D.A.-H.) Investigation, Validation, Formal analysis, Writing, Original Draft, Visualization. (E.J.O.-R.) Investigation, Formal analysis. (D.L.M.-Z.) Investigation, Visualization. (E.M.-N.) Conceptualization, Methodology, Validation, Resources, Writing, Review and Editing, Visualization, Supervision, Project administration, funding acquisition. All authors have given approval to the final version of the manuscript.
Funding
This work was supported by SICES (Guanajuato, Mexico, Grant Nos. CMTEVAL-18-S01-03 and SICES/CONV/252/2019) and CONACYT (Mexico, Grant No. 312271, Apoyo para Proyectos de Investigación Científica, Desarrollo Tecnológico e Innovación en Salud ante la Contingencia por COVID-19; Grant No. 299058, Laboratorio Nacional de Micro y Nanofluídica; Grant No. 297497, FORDECYT, “Generación de Plataformas tecnológicas basadas en Microdispositivos para el sector Industrial de los estados de Aguascalientes, Guanajuato, Puebla, Querétaro y San Luis Potosı”).
Acknowledgments
The authors are grateful to La Próxima Estación S.P.R. de R.L.
References
- Majdinasab, M.; Hayat, A.; Marty, J.L. Aptamer-based assays and aptasensors for detection of pathogenic bacteria in food samples. TrAC Trends Anal. Chem. 2018, 107, 60–77. [Google Scholar] [CrossRef]
- Wang, Y.; Duncan, T.V. Nanoscale sensors for assuring the safety of food products. Food Biotechnol. Plant Biotechnol. 2017, 44, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Avila-Huerta, M.D.; Ortiz-Riaño, E.J.; Mancera-Zapata, D.L.; Morales-Narváez, E. Real-Time Photoluminescent Biosensing Based on Graphene Oxide-Coated Microplates: A Rapid Pathogen Detection Platform. Anal. Chem. 2020, 92, 11511–11515. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Bhardwaj, S.K.; Bhatt, D.; Lim, D.K.; Kim, K.H.; Deep, A. Optical detection of waterborne pathogens using nanomaterials. TrAC Trends Anal. Chem. 2019, 113, 280–300. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Nayak, M.K.; Mehta, J.; Kim, K.H.; Deep, A. Fluorescent nanobiosensors for the targeted detection of foodborne bacteria. TrAC Trends Anal. Chem. 2017, 97, 120–135. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, N.; Kumari, A.; Lee, H.J.; Kim, T.; Tripathi, K.M. Nano-carbon based sensors for bacterial detection and discrimination in clinical diagnosis: A junction between material science and biology. Appl. Mater. Today 2020, 18, 100467. [Google Scholar] [CrossRef]
- Luo, K.; Kim, H.-Y.; Oh, M.-H.; Kim, Y.-R. Paper-based lateral flow strip assay for the detection of foodborne pathogens: Principles, applications, technological challenges and opportunities. Crit. Rev. Food Sci. Nutr. 2018, 60, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-Y.; Hulme, J.; An, S.S.A. Recent trends in the detection of pathogenic Escherichia coli O157: H7. BioChip J. 2015, 9, 173–181. [Google Scholar] [CrossRef]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Riaño, E.J.; Avila-Huerta, M.D.; Mancera-Zapata, D.L.; Morales-Narváez, E. Microwell plates coated with graphene oxide enable advantageous real-time immunosensing platform. Biosens. Bioelectron. 2020, 165, 112319. [Google Scholar] [CrossRef] [PubMed]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).