Electrokinetically Assisted Paper-Based DNA Concentration for Enhanced qPCR Sensing †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Device Fabrication
2.2. Electrophoresis
2.3. qPCR
3. Results and Discussion
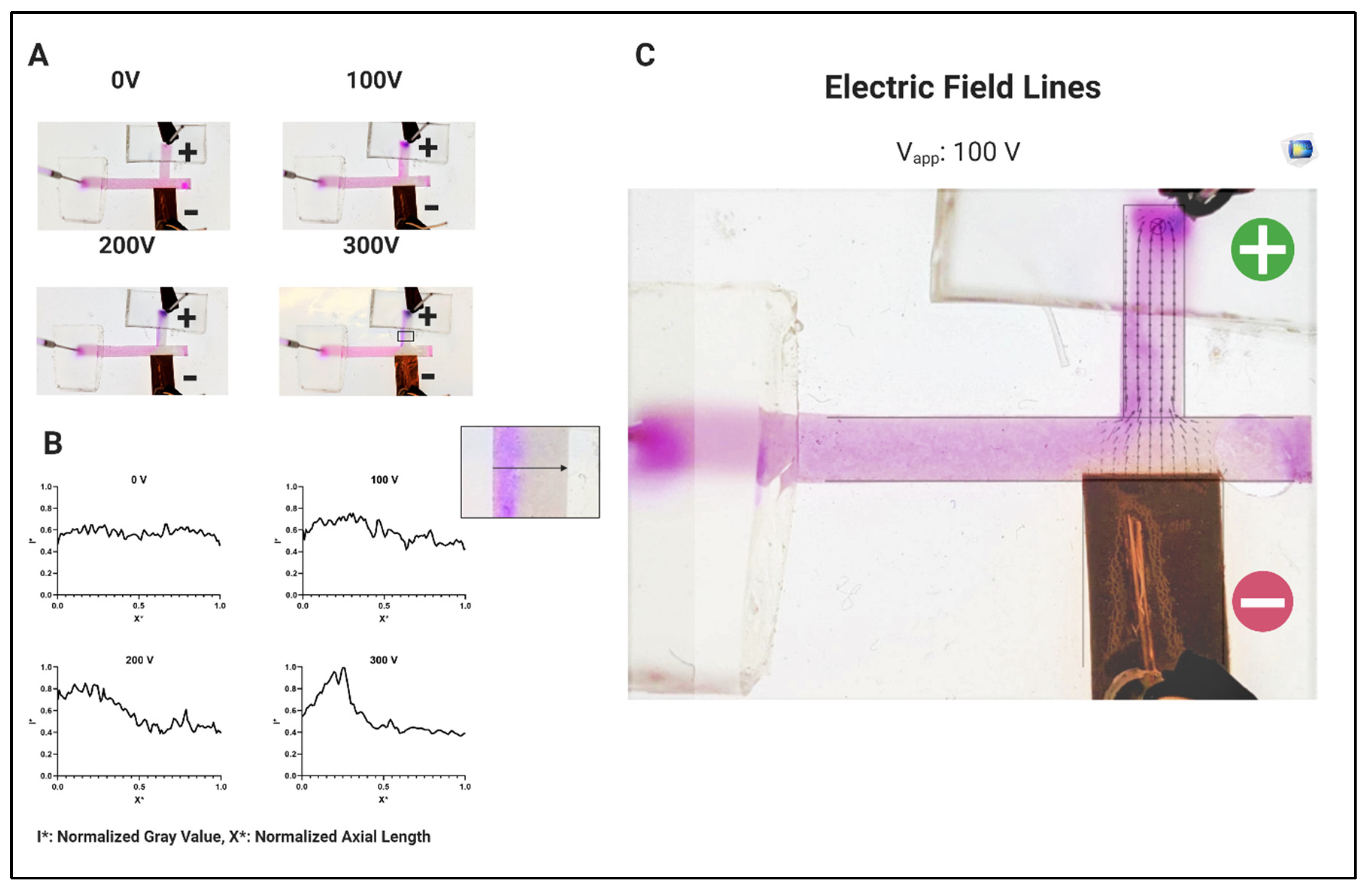
3.1. System Characterization Using Charged Dyes
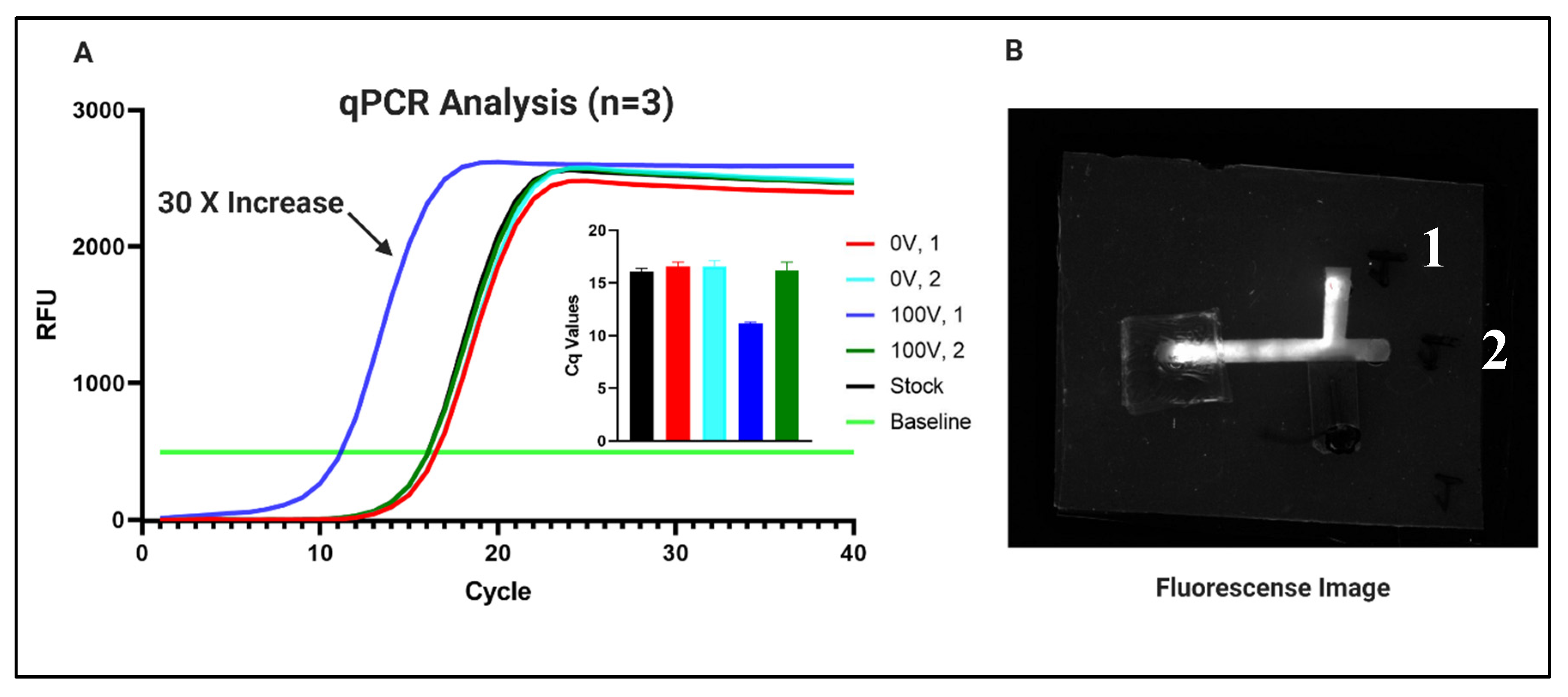
3.2. Nucleic Acid Concentration
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Convery, N.; Gadegaard, N. 30 years of microfluidics. Micro Nano Eng. 2019, 2, 76–91. [Google Scholar] [CrossRef]
- Microfluidics Market Size, Share & Trends Analysis Report By Application (Lab-on-a-Chip, Organs-on-Chips, Continuous Flow), By Technology (Medical, Non-Medical), By Material, And Segment Forecasts, 2020–2027; Grand View Research: San Francisco, CA, USA, 2020.
- Faustino, V.; Catarino, S.O.; Lima, R.; Minas, G. Biomedical microfluidic devices by using low-cost fabrication techniques: A review. J. Biomech. 2016, 49, 2280–2292. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.I., 3rd; Kong, D.S.; Murthy, S.K.; Carr, P.A. Enabling Microfluidics: From Clean Rooms to Makerspaces. Trends Biotechnol. 2017, 35, 383–392. [Google Scholar] [CrossRef]
- Gale, B.; Jafek, A.; Lambert, C.; Goenner, B.; Moghimifam, H.; Nze, U.; Kamarapu, S. A Review of Current Methods in Microfluidic Device Fabrication and Future Commercialization Prospects. Inventions 2018, 3, 60. [Google Scholar] [CrossRef]
- Carrell, C.; Kava, A.; Nguyen, M.; Menger, R.; Munshi, Z.; Call, Z.; Nussbaum, M.; Henry, C. Beyond the lateral flow assay: A review of paper-based microfluidics. Microelectron. Eng. 2019, 206, 45–54. [Google Scholar] [CrossRef]
- Martinez, A.W.; Phillips, S.T.; Butte, M.J.; Whitesides, G.M. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew. Chem. Int. Ed. Engl. 2007, 46, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Sher, M.; Zhuang, R.; Demirci, U.; Asghar, W. Paper-based analytical devices for clinical diagnosis: Recent advances in the fabrication techniques and sensing mechanisms. Expert Rev. Mol. Diagn. 2017, 17, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Soum, V.; Park, S.; Brilian, A.I.; Kwon, O.S.; Shin, K. Programmable Paper-Based Microfluidic Devices for Biomarker Detections. Micromachines (Basel) 2019, 10, 516. [Google Scholar] [CrossRef] [PubMed]
- Osborn, J.L.; Lutz, B.; Fu, E.; Kauffman, P.; Stevens, D.Y.; Yager, P. Microfluidics without pumps: Reinventing the T-sensor and H-filter in paper networks. Lab Chip 2010, 10, 2659–2665. [Google Scholar] [CrossRef] [PubMed]
- Määttänen, A.; Fors, D.; Wang, S.; Valtakari, D.; Ihalainen, P.; Peltonen, J. Paper-based planar reaction arrays for printed diagnostics. Sens. Actuators B Chem. 2011, 160, 1404–1412. [Google Scholar] [CrossRef]
- Islam, M.N.; Ahmed, I.; Anik, M.I.; Ferdous, M.S.; Khan, M.S. Developing Paper Based Diagnostic Technique to Detect Uric Acid in Urine. Front. Chem. 2018, 6, 496. [Google Scholar] [CrossRef] [PubMed]
- Andres, W.; Martinez, S.T.P.; Carrilho, E.; Whitesides, G.M. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal. Chem. 2010, 82, 3–10. [Google Scholar]
- Cheng, C.M.; Martinez, A.W.; Gong, J.; Mace, C.R.; Phillips, S.T.; Carrilho, E.; Mirica, K.A.; Whitesides, G.M. Paper-based ELISA. Angew. Chem. Int. Ed. Engl. 2010, 49, 4771–4774. [Google Scholar] [CrossRef] [PubMed]
- Jokerst, J.C.; Adkins, J.A.; Bisha, B.; Mentele, M.M.; Goodridge, L.D.; Henry, C.S. Development of a paper-based analytical device for colorimetric detection of select foodborne pathogens. Anal. Chem. 2012, 84, 2900–2907. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.N.; Yost, J.; Gagnon, Z. Electrokinetically Assisted Paper-Based DNA Concentration for Enhanced qPCR Sensing. Proceedings 2020, 60, 33. https://doi.org/10.3390/IECB2020-07074
Islam MN, Yost J, Gagnon Z. Electrokinetically Assisted Paper-Based DNA Concentration for Enhanced qPCR Sensing. Proceedings. 2020; 60(1):33. https://doi.org/10.3390/IECB2020-07074
Chicago/Turabian StyleIslam, Md Nazibul, Jarad Yost, and Zachary Gagnon. 2020. "Electrokinetically Assisted Paper-Based DNA Concentration for Enhanced qPCR Sensing" Proceedings 60, no. 1: 33. https://doi.org/10.3390/IECB2020-07074
APA StyleIslam, M. N., Yost, J., & Gagnon, Z. (2020). Electrokinetically Assisted Paper-Based DNA Concentration for Enhanced qPCR Sensing. Proceedings, 60(1), 33. https://doi.org/10.3390/IECB2020-07074



