Web Server and R Library for the Calculation of Markov Chains Molecular Descriptors †
Abstract
:1. Introduction
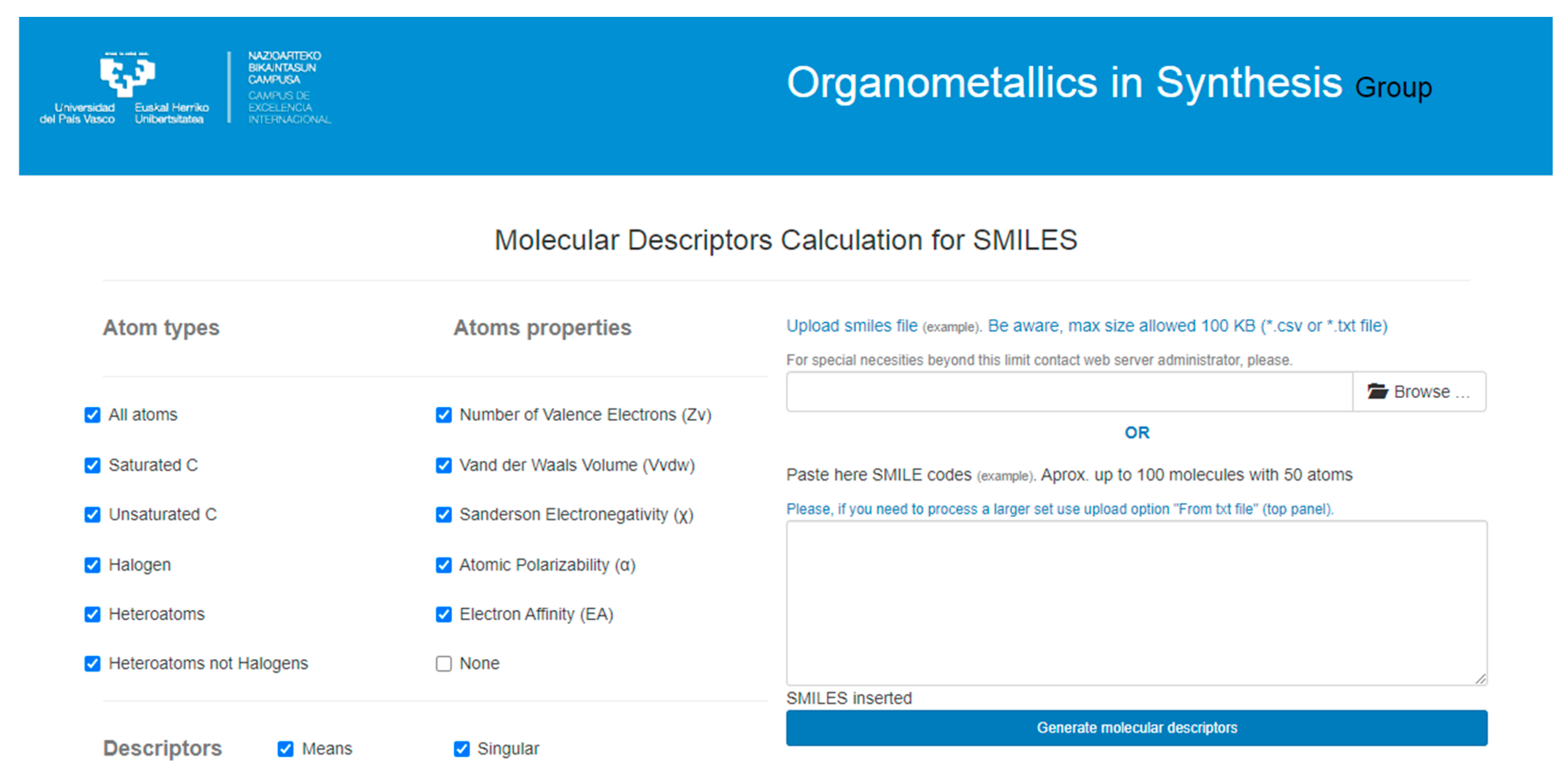
2. Materials and Methods
3. Results
4. Conclusions
References
- Riihimaki, M.; Hemminki, A.; Sundquist, J.; Hemminki, K. Pat-terns of Metastasis in Colon and Rectal Cancer. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jhanwar, B.; Sharma, V.; Singla, R.K.; Shrivastava, B. QSAR-Hansch Analysis and Related Approaches in Drug Design. Phar.-Macol. Online Newsl. 2011, 1, 306–344. [Google Scholar]
- Santiago, C.B.; Guo, J.Y.; Sigman, M.S. Predictive and Mechanistic Multivariate Linear Regression Models for Reaction Development. Chem. Sci. 2018, 9, 2398–2412. [Google Scholar] [CrossRef] [PubMed]
- Tsiliki, G.; Munteanu, C.R.; Seoane, J.A.; Fernandez-Lozano, C.; Sarimveis, H.; Willighagen, E.L. RRegrs: An R package for computer-aided model selection with multiple regression models. J. Cheminform. 2015, 7, 46. [Google Scholar] [CrossRef] [PubMed]
| Method | Training | Test | ||
|---|---|---|---|---|
| R2 | RMSE | R2 | RMSE | |
| RF | 0.907 | 0.101 | 0.926 | 0.093 |
| SVM | 0.868 | 0.122 | 0.866 | 0.128 |
| NN | 0.849 | 0.132 | 0.829 | 0.143 |
| PLS | 0.801 | 0.155 | 0.775 | 0.167 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carracedo-Reboredo, P.; Munteanu, C.R.; González-Díaz, H.; Fernandez-Lozano, C. Web Server and R Library for the Calculation of Markov Chains Molecular Descriptors. Proceedings 2020, 54, 28. https://doi.org/10.3390/proceedings2020054028
Carracedo-Reboredo P, Munteanu CR, González-Díaz H, Fernandez-Lozano C. Web Server and R Library for the Calculation of Markov Chains Molecular Descriptors. Proceedings. 2020; 54(1):28. https://doi.org/10.3390/proceedings2020054028
Chicago/Turabian StyleCarracedo-Reboredo, Paula, Cristian R. Munteanu, Humbert González-Díaz, and Carlos Fernandez-Lozano. 2020. "Web Server and R Library for the Calculation of Markov Chains Molecular Descriptors" Proceedings 54, no. 1: 28. https://doi.org/10.3390/proceedings2020054028
APA StyleCarracedo-Reboredo, P., Munteanu, C. R., González-Díaz, H., & Fernandez-Lozano, C. (2020). Web Server and R Library for the Calculation of Markov Chains Molecular Descriptors. Proceedings, 54(1), 28. https://doi.org/10.3390/proceedings2020054028








