Intracellular Background Estimation for Quantitative Fluorescence Microscopy †
Abstract
:1. Introduction
2. Two Background Level Estimation (TBL) Algorithm
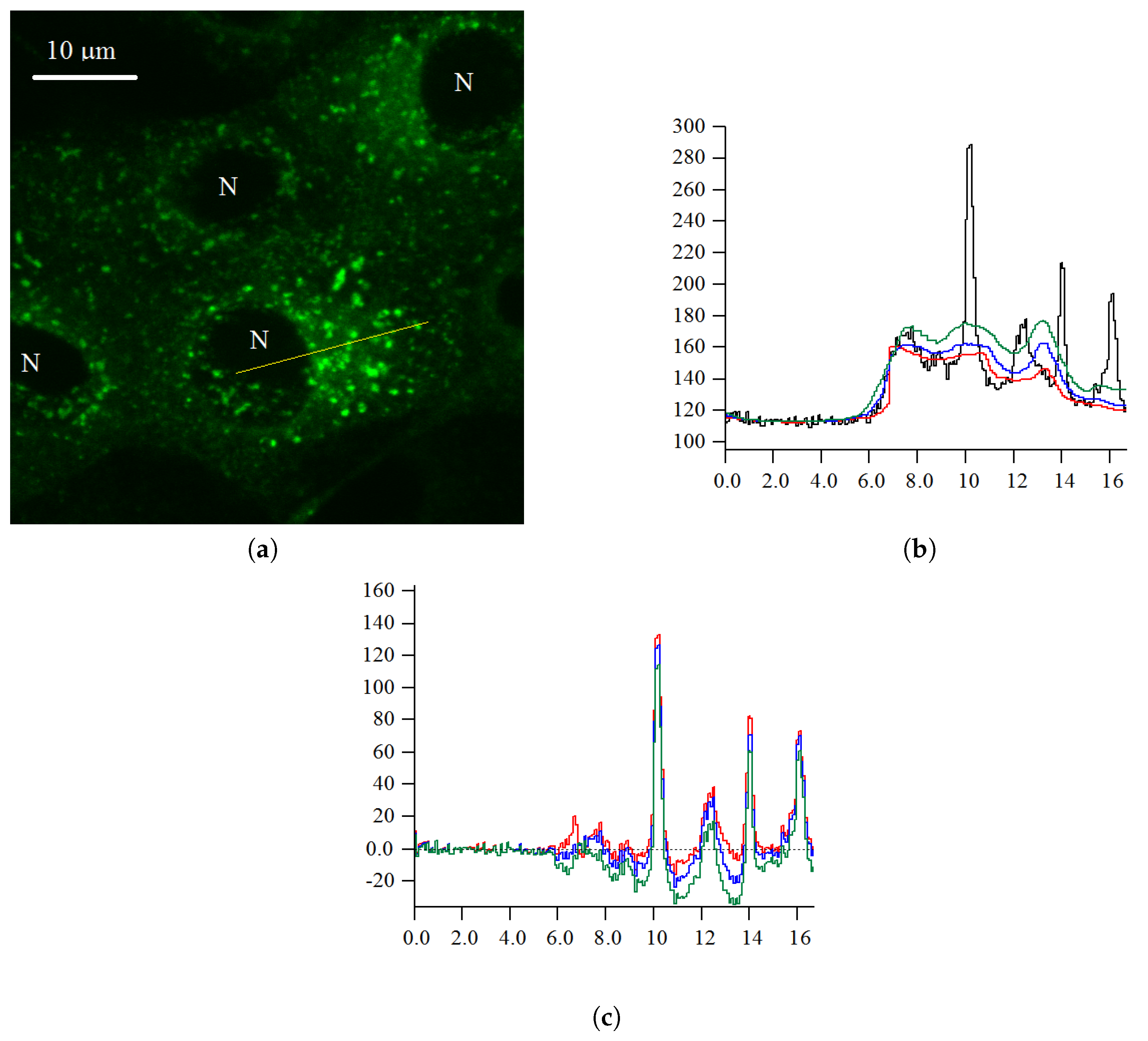
2.1. Probabilistic Model of Intensity
2.2. Probabilistic Model of Two Background Levels
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Estimation Prior Parameters μ and β
References
- Zimmer, M. Green Fluorescent Protein (GFP): Applications, Structure, and Related Photophysical Behavior. Chem. Rev. 2002, 102, 759–782. [Google Scholar] [CrossRef] [PubMed]
- Rink, J.C.; Ghigo, E.; Kalaidzidis, Y.L.; Zerial, M. Rab Conversion as a Mechanism of Progression from Early to Late Endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E.; Smal, I.; Danuser, G. Tracking in Molecular Bioimaging. IEEE Signal Process. Mag. 2006, 23, 46–53. [Google Scholar] [CrossRef]
- Sbalzarini, I.F.; Koumoutsakos, P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2006, 151, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R. Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol. Biol. Cell 2017, 28, 712–715. [Google Scholar] [CrossRef] [PubMed]
- Pécot, T.; Bouthemy, P.; Boulanger, J.; Chessel, A.; Bardin, S.; Salamero, J.; Kervrann, C. Background Fluorescence Estimation and Vesicle Segmentation in Live Cell Imaging with Conditional Random Fields. IEEE Trans. Image Process. 2015, 24, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Kalaidzidis, Y. (Version 8.93.00, 27 June 2019). Available online: http://motiontracking.mpi-cbg.de.
- Lee, H.-C.; Yang, G. Computational Removal of Background Fluorescence for Biological Fluorescence Microscopy. In Proceedings of the IEEE 11th International Symposium on Biomedical Imaging (ISBI), Beijing, China, 29 April–2 May 2014. [Google Scholar]
- Yang, L.; Zhang, Y.; Guldner, I.H.; Zhang, S.; Chen, D.Z. Fast Background Removal in 3D Fluorescence Microscopy Images Using One-Class Learning. In International Conference on Medical Image Computing and Computer-Assisted Intervention—ICCAI 2015, Part III; Springer: Cham, Switzerland, 2015; pp. 292–299. [Google Scholar]
- Sternberg, S.R. Biomedical Image Processing. Computer (IEEE) 1983, 16, 22–34. [Google Scholar] [CrossRef]
- Kalaidzidis, Y. Fluorescence Microscopy Noise Model: Estimation of Poisson Noise Parameters from Snap-Shot Image. In Proceedings of the International Conference on Bioinformatics and Computational Biology (BIOCOMP’17), Las Vegas, NV, USA, 17–20 July 2017; pp. 63–66. [Google Scholar]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalaidzidis, Y.; Morales-Navarrete, H.; Kalaidzidis, I.; Zerial, M. Intracellular Background Estimation for Quantitative Fluorescence Microscopy. Proceedings 2019, 33, 22. https://doi.org/10.3390/proceedings2019033022
Kalaidzidis Y, Morales-Navarrete H, Kalaidzidis I, Zerial M. Intracellular Background Estimation for Quantitative Fluorescence Microscopy. Proceedings. 2019; 33(1):22. https://doi.org/10.3390/proceedings2019033022
Chicago/Turabian StyleKalaidzidis, Yannis, Hernán Morales-Navarrete, Inna Kalaidzidis, and Marino Zerial. 2019. "Intracellular Background Estimation for Quantitative Fluorescence Microscopy" Proceedings 33, no. 1: 22. https://doi.org/10.3390/proceedings2019033022
APA StyleKalaidzidis, Y., Morales-Navarrete, H., Kalaidzidis, I., & Zerial, M. (2019). Intracellular Background Estimation for Quantitative Fluorescence Microscopy. Proceedings, 33(1), 22. https://doi.org/10.3390/proceedings2019033022





