Standalone Point-of-Use Device for Gluten Detection in Food: POCT Application Experiment in SMARTER-SI European Project †
Abstract
:1. Introduction
2. Antigens and Antibodies
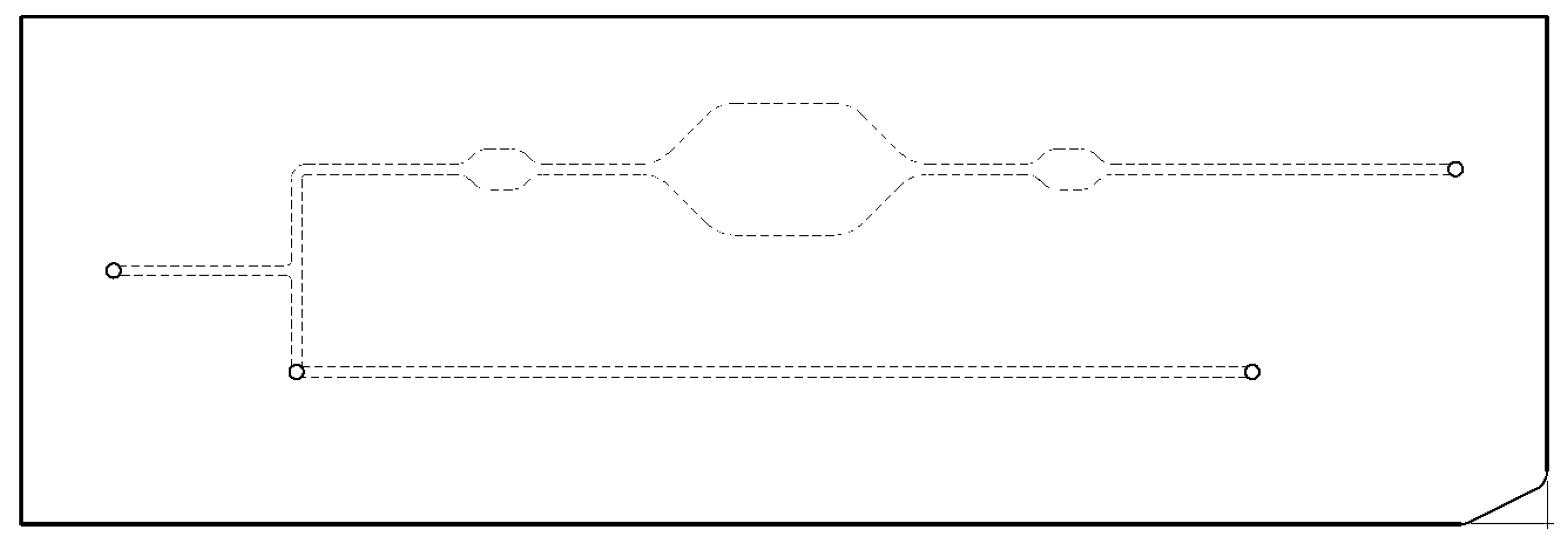
3. Microfluidic Cartridges
4. Antigen Immobilization
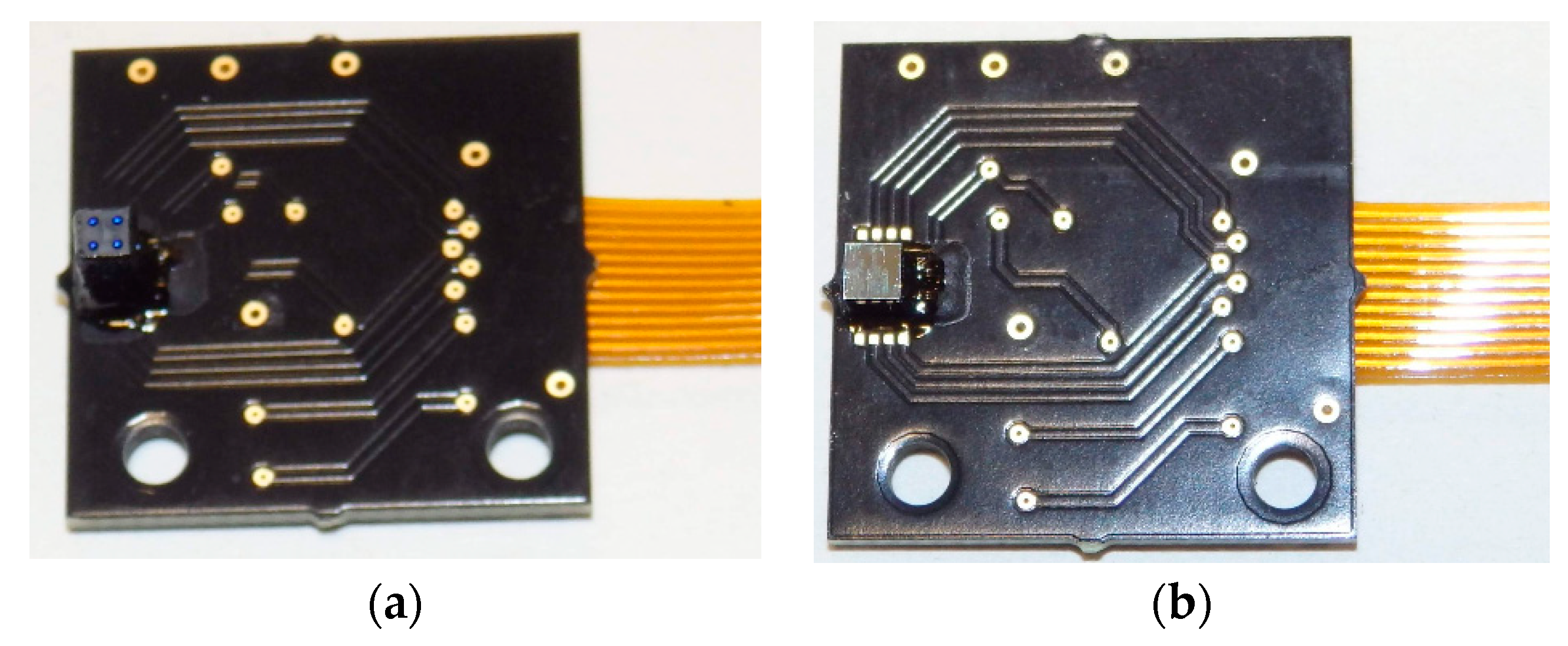
5. Microfluidic Microarray Immunoassay
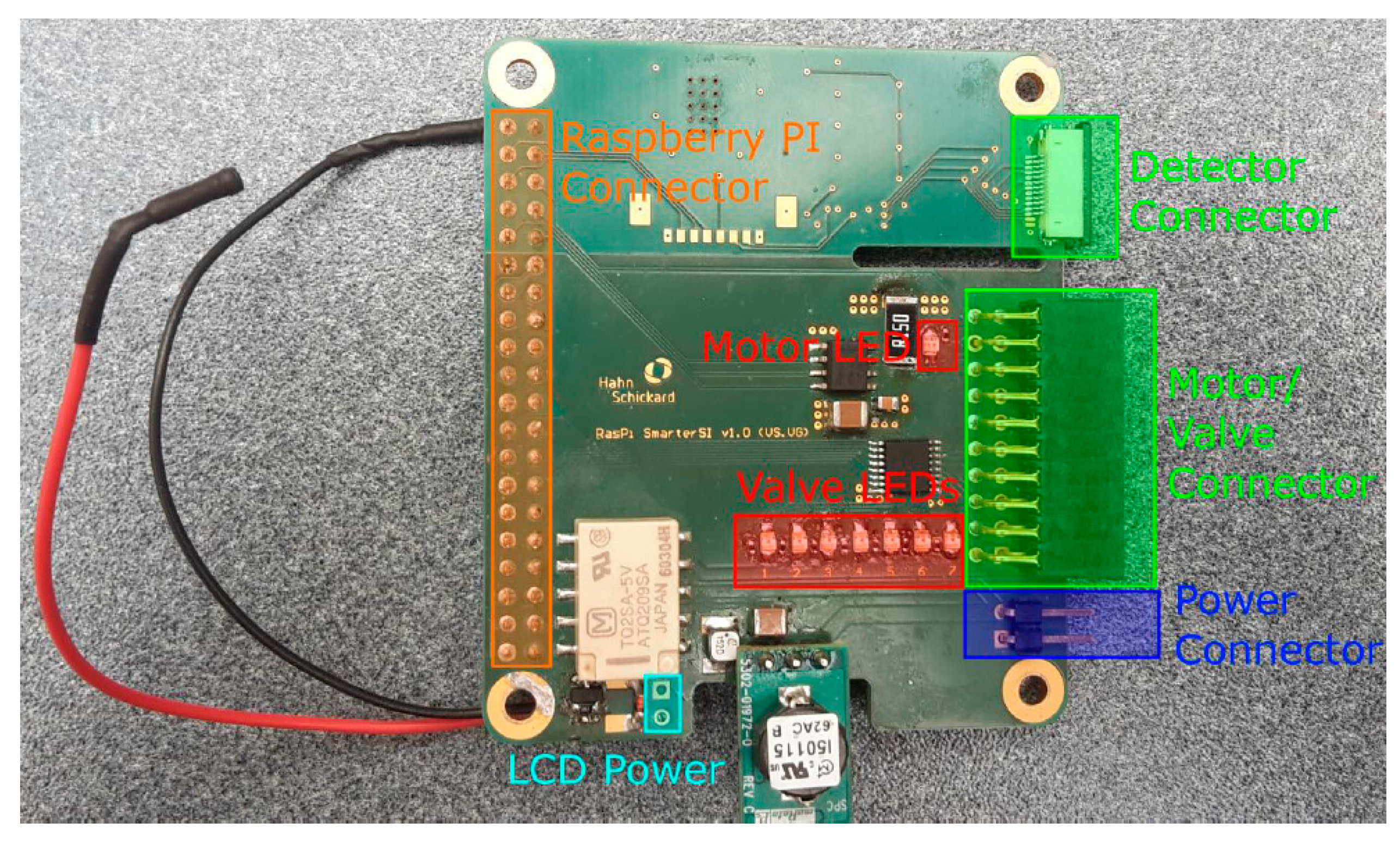
6. Platform
6.1. Colorimetric Version
6.2. Fluorescence Version
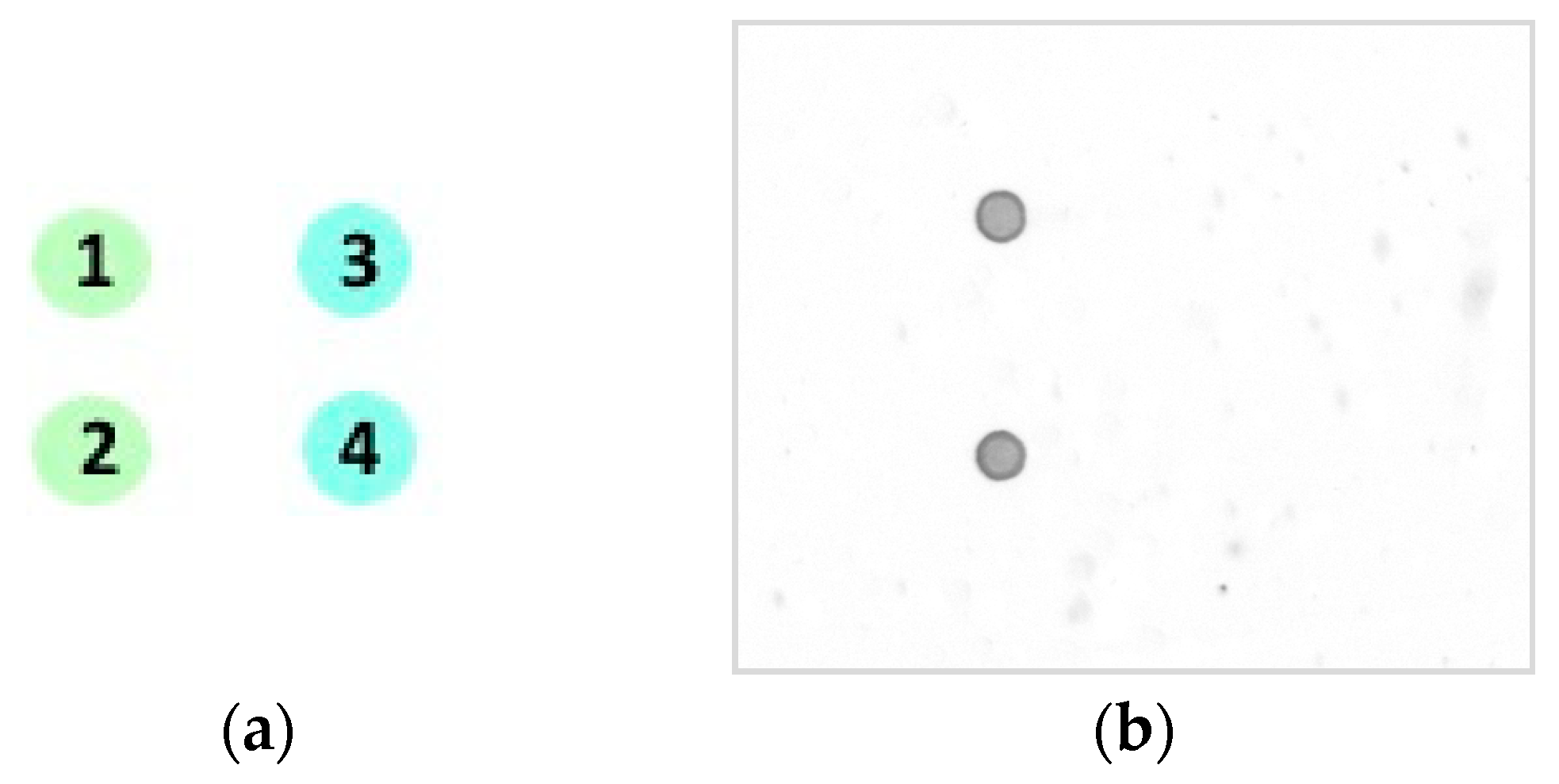
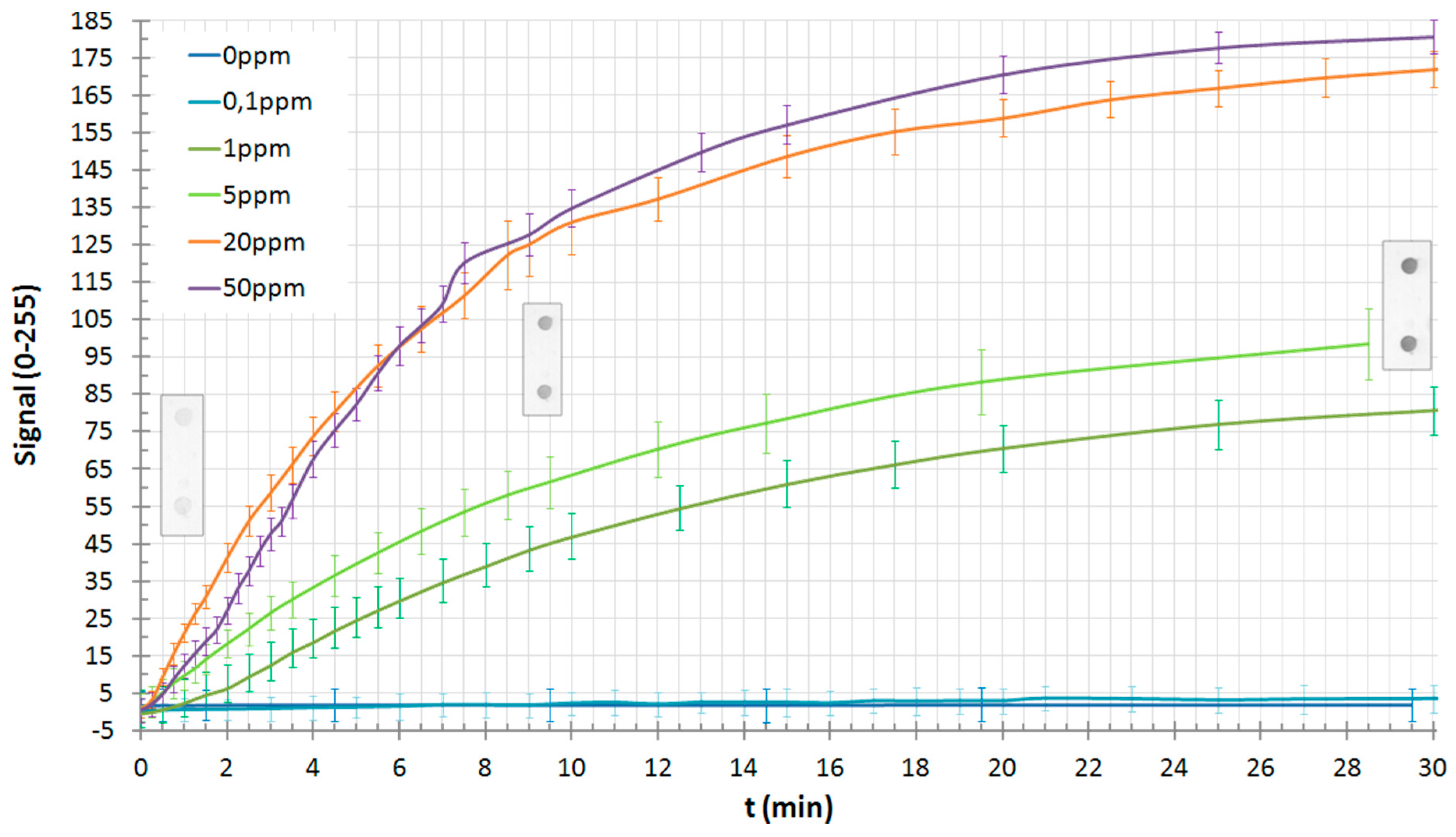
7. Results
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ursula, S. Analytical protein microarrays: Advancements towards clinical applications. Sensors 2017, 17, 256. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Mustalahti, K.; Catassi, C.; Reunanen, A.; Fabiani, E.; Heier, M.; McMillan, S.; Murray, L.; Metzger, M.H.; Gasparin, M.; Bravi, E.; et al. The prevalence of celiac disease in Europe: Results of a centralized, international mass screening project. Ann. Med. 2010, 42, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Peter, H.R.; Green, M.D.; Christophe Cellier, M.D. Celiac disease. N. Engl. J. Med. 2007, 357, 1731–1743. [Google Scholar]
- Codex Alimentarius, CODEX STAN 118-1979. Available online: http://www.codexalimentarius.org/ (accessed on 2 October 2017).
- Coeliac Disease New Zealand. Available online: http://www.coeliac.org.nz/food-industry/gluten-free-food-standards (accessed on 2 October 2017).
- Valdés, I.; García, E.; Llorente, M.; Méndez, E. Innovative approach to low-level gluten determination in foods using a novel sandwich enzyme-linked immunosorbent assay protocol. Eur. J. Gastroenterol. Hepatol. 2003, 15, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Nakane, P.K.; Kawaoi, A. Peroxidase-labeled antibody. A new method of conjugation. J. Histochem. Cytochem. 1974, 22, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Elizalde, J.; Mayora, K.; Rivas, L.A.; Sanz, A.; Tolentino-Cortez, T.; Barreda-Gómez, G.; Tijero, M. Multi-parametric point of care device for allergen-specific IgE detection in veterinary applications. Procedia Eng. 2016, 168, 1410–1413. [Google Scholar] [CrossRef]
- Raspberry Pi Foundation. Available online: https://www.raspberrypi.org/ (accessed on 2 October 2017).
- Python Software Foundation. Python Language Reference, Version 2.7. Available online: https://docs.python.org/2/ (accessed on 2 October 2017).
- Kivy Organization. Available online: https://kivy.org/ (accessed on 2 October 2017).
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elizalde, J.; Eguiluz, X.; Mayora, K.; Rivas, L.A.; Sanz, A.; Dehnert, J.; Goridko, V.; Görlandt, S.; Schädel, M. Standalone Point-of-Use Device for Gluten Detection in Food: POCT Application Experiment in SMARTER-SI European Project. Proceedings 2018, 2, 128. https://doi.org/10.3390/ecsa-4-04915
Elizalde J, Eguiluz X, Mayora K, Rivas LA, Sanz A, Dehnert J, Goridko V, Görlandt S, Schädel M. Standalone Point-of-Use Device for Gluten Detection in Food: POCT Application Experiment in SMARTER-SI European Project. Proceedings. 2018; 2(3):128. https://doi.org/10.3390/ecsa-4-04915
Chicago/Turabian StyleElizalde, Jorge, Xabier Eguiluz, Kepa Mayora, Luis Alfonso Rivas, Antonio Sanz, Jan Dehnert, Vadim Goridko, Stefan Görlandt, and Martin Schädel. 2018. "Standalone Point-of-Use Device for Gluten Detection in Food: POCT Application Experiment in SMARTER-SI European Project" Proceedings 2, no. 3: 128. https://doi.org/10.3390/ecsa-4-04915
APA StyleElizalde, J., Eguiluz, X., Mayora, K., Rivas, L. A., Sanz, A., Dehnert, J., Goridko, V., Görlandt, S., & Schädel, M. (2018). Standalone Point-of-Use Device for Gluten Detection in Food: POCT Application Experiment in SMARTER-SI European Project. Proceedings, 2(3), 128. https://doi.org/10.3390/ecsa-4-04915




