Abstract
In both healthy cells and cancer cells a number of different cellular responses influence the progress of cancer. In this study, we have investigated the effect of stress conditions on human hepatocyte cancer cells. Different concentrations of hydrogen peroxide, quercetin and N-acetyl cysteine were used to induce the acute cell stress. We have detected apoptosis, measured proteasome activity and performed a cell death array in both treated and control cells. Treatment of human hepatocyte cancer cells with hydrogen peroxide and quercetin resulted in decreased cell viability and increased apoptosis. On the other hand, proteasome activity was increased with hydrogen peroxide treatment but decreased with quercetin. N-acetyl cysteine supported cell viability and increased proteasome activity. In conclusion, cell death related genes were affected in quite different manner following hydrogen peroxide, quercetin and N-acetyl cysteine treatments.
1. Introduction
Hepatocellular carcinoma causes over 600,000 deaths per year and is the most common type of liver tumors. It is also difficult to treat because of the resistance to current chemotherapies [1]. Healthy cells and cancer cells influence the progress of cancer through different types of responses. A number of different stress response mechanisms are involved in the progress and therapy resistance of hepatocellular carcinoma (HepG2) cells. Stress is defined as any disturbance of organism’s homeostasis and is a ubiquitous phenomenon shaping the life throughout evolution. Adaptation to living in a stressful environment is possible through establishment of numerous stress response mechanisms. Although multicellularity, humoral regulations and thermoregulation eliminate a host of direct stresses to human cells, proper stress response at the cellular level is still critical for the survival of organisms [2]. In this study, we have investigated the effect of stress conditions on human HepG2 cells. Hydrogen peroxide, and quercetin were used to induce acute cell stress and N-acetylcysteine was used as positive control. We have evaluated apoptosis, measured proteasome activity and performed a cell death array in both treated and control cells.
2. Materials and Methods
In this study hydrogen peroxide and quercetin were used to induce acute stress, and N-acetylcysteine was used as positive control. Cells were cultured in DMEM supplemented with 10% fetal bovine serum, 1% l-glutamine and 1% penicillin/streptomycin in a 5% CO2 incubator at 37 °C. Cell viability was monitored by WST1 assay. For detection of apoptosis, 0.5 × 106 cells were washed with PBS and ApopNexin FITC detection kit was used in FACSCalibur flow cytometry system. All experiments were performed in triplicate and in each assay 1 × 104 cells were measured. Results were assessed using the CellQuest program. Proteasome activity was measured with the fluorogenic peptide succinyl-leucine-leucine-valine-tyrosine-methylcoumarin at a concentration of 200 µM. Fluorescence was measured at 360 nm excitation and 485 nm emission. Proteasome activity was calculated using free methylcoumarin as standard. RNA isolation from cells was performed by NORGEN Total RNA isolation kit. Following isolation of RNA, the Human Cell Death Pathway Finder RT2 Profiler PCR Array was used to evaluate the expression of 84 key genes which are involved in the central mechanisms of cellular death: apoptosis, autophagy, and necrosis.
3. Results
HepG2 cells were treated with different concentrations of hydrogen peroxide, quercetin and N-acetyl cysteine for 30 min to investigate the effects of acute stress. As shown in Figure 1, viability of HepG2 cells was decreased with hydrogen peroxide and quercetin treatment, and was increased with N-acetyl cysteine. Percentage of total apoptotic cells were 9.66 for 50 µM hydrogen peroxide, 10.93 for 50 µM quercetin, 16.45 for 200 µM hydrogen peroxide and 18.78 for 200 µM quercetin.
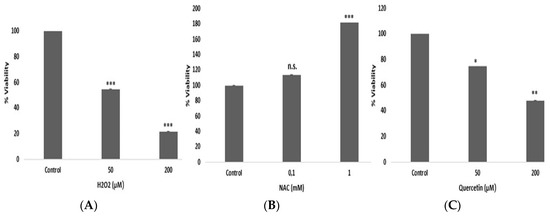
Figure 1.
Percentage viability of HepG2 cells treated with H2O2 (A), N-acetyl cysteine (B), and quercetin (C). * p < 0.05 vs. control; ** p < 0.01 vs. control; *** p < 0.001 vs. control cells.
Proteasome activity was measured in -ATP, ATP-depleted and +ATP conditions. Quercetin decreased proteasome activity significantly (data not shown). We have analyzed the expression of 84 genes in hydrogen peroxide, quercetin and N-acetyl cysteine treated cells for characterization of stress dependent transcriptional effects. These genes were involved in pro-, anti-apoptotic, autophagy and necrosis pathways. In general, thirty minutes of stress resulted in down-regulation of most of the observed genes but only several genes were significantly changed. In hydrogen peroxide treated cells 10 genes were observed to change, only six of them were statistically significant. These genes were heat shock 27 associated protein 1 (HSPBAP1), myelin associated glycoprotein (MAG), olfactory receptor family 10 sub family J member 3 (OR10J3), synaptonemal complex protein 2 (SYCP2), interferon gamma (IFNG), insulin-like growth factor-1 (IGF1), insulin (INS), immunity related GTPase family M (IRGM), synuclein alpha (nonA3 complement of amyloid precursor) (SNCA) and tumor necrosis factor receptor super family member 11b (TNFRSF11B). Quercetin changed the expression of 26 genes and 16 of them were significantly changed. Five of these genes were pro-apoptotic, one of them was anti-apoptotic, six of them were necrotic and four of them were associated autophagy. These were autophagy related 16-like 1 (ATG16L1), BCL2-related protein A1 (BCL2A1), BCL2-like 1 (BCL2L1), BCL2-like 11 (BCL2L11), caspase 7 (CASP7), coiled-coil domain containing 103 (CCDC103), CD40, N-acetyl galactosaminyl-transferase 5 (GALNT5), growth factor receptor-bound protein 2 (GRB2), interferon gamma (IFNG), insulin (INS), myelin associated glycoprotein (MAG), RAB25 member RAS oncogene family (RAB25), synaptonemal complex protein 2 (SYCP2), tumor necrosis factor receptor superfamily member 1A (TNFRSF1A). Several genes involved in regulation of cell death process were down-regulated by NAC, 19 of these genes were changed and 6 of them were significant.
4. Discussion
Previous reports showed that quercetin is cytotoxic in various human cancer cell lines with varying sensitivity [3,4,5] and varying range of IC50 was reported for quercetin from 10 to 100 μM. K562 cells, a chronic myelogenous leukemia cell line, was reported to display resistance to most of the anti-cancerous drugs. However, in previous studies it was also observed to be sensitive towards quercetin [6]. Our results in HepG2 cells showed that quercetin could significantly inhibit proliferation and induce apoptosis via inhibition of proteasome activity. Quercetin concentrations that significantly decrease HepG2 cell viability have similar effects on other cancer cells such as leukemia [7]. Our result with quercetin further demonstrates the inhibitory effect of this flavonoid on tumor cell viability. Expression of BCL2A1, BCL2L1, and BCL2L11 could be differently regulated by quercetin suggesting that the balance in the expression of these proteins might be involved in the control of the apoptotic process. Hydrogen peroxide treatment increased the expression of one autophagy and one necrosis gene while decreasing three autophagy genes and one necrosis gene. We observed that quercetin, a natural polyphenol, can act as a potent apoptotic compound restraining necrosis and autophagy activation, and contributing to modulation of apoptosis. We also found that IGF1 gene levels were up-regulated following hydrogen peroxide and quercetin. This gene is associated with autophagy in the cell death pathway. IGF1 is a crucial ligand for IGF pathway, it is highly expressed and known to promote the growth of several tumors. This is not the case in liver cancer, where it has been strongly suggested as an anti-tumorigenic factor. Another up-regulated gene was TNF, which is identified as a pro-apoptotic gene in cell death pathway. TNF is characterized by high malignancy recurrence and short survival time. Inflammation is closely related to the initiation and development of hepatocellular carcinoma. Previous studies showed that TNF expression was increased in both human tumor tissues and cell lines (HepG2 and HuH7) and high TNF expression was associated with hepatocellular carcinoma development [8]. ATG6V1G2 gene, which is involved in autophagy pathway, was up-regulated following quercetin treatment. ATG6V1G2 gene is associated with intracellular processes such as receptor mediated endocytosis and synaptic vesicle proton gradient generation. In conclusion, future direction of our study will be to analyze corresponding protein levels of these genes and to test the same pathways in exosomes released from cancer cells.
Acknowledgments
This study was supported by The Scientific and Technological Research Council of Turkey (TÜBİTAK, Project No. 216S963).
References
- Nakanishi, K.; Sakamoto, M.; Yamasaki, S.; Todo, S.; Hirohashi, S. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer 2005, 103, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Zelenka, J.; Koncosova, M.; Ruml, T. Targeting of stress response pathways in the prevention and treatment of cancer. Biotechnol. Adv. 2018, 36, 583–602. [Google Scholar] [CrossRef] [PubMed]
- Mertens-Talcott, S.U.; Percival, S.S. Ellagic acid and quercetin interact synergistically with resveratrol in the induction of apoptosis and cause transient cell cycle arrest in human leukemia cells. Cancer Lett. 2005, 218, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Borska, S.; Chmielewska, M.; Wysocka, T.; Drag-Zalesinska, M.; Zabel, M.; Dziegiel, P. In vitro effect of quercetin on human gastric carcinoma: Targeting cancer cells death and MDR. Food Chem. Toxicol. 2012, 50, 3375–3383. [Google Scholar] [CrossRef]
- Wenzel, U.; Herzog, A.; Kuntz, S.; Daniel, H. Protein expression profiling identifies molecular targets of quercetin as a major dietary flavonoid in human colon cancer cells. Proteomics 2004, 4, 2160–2174. [Google Scholar] [CrossRef]
- Brisdelli, F.; Coccia, C.; Cinque, B.; Cifone, M.G.; Bozzi, A. Induction of apoptosis by quercetin: Different response of human chronic myeloid (K562) and acute lymphoblastic (HSB-2) leukemia cells. Mol. Cell. Biochem. 2007, 296, 137–149. [Google Scholar] [CrossRef]
- Mutlu Altundag, E.; Yılmaz, A.M.; Kocturk, S.; Taga, Y.; Yalcın, A.S. Synergistic induction of apoptosis by quercetin and curcumin in chronic myeloid leukemia (K562) cells. Nutr. Cancer 2018, 70, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, J.; Hu, X.; Liu, S.; He, B. Prognostic and therapeutic values of tumor necrosis factor alpha in hepatocellular carcinoma. Med. Sci. Monit. 2016, 22, 3694–3704. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).