Biodegradation of 5-(Hydroxymethyl)-furfural and Furan Derivatives †
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Acknowledgments
Conflicts of Interest
References
- Barakat, A.; Monlau, F.; Steyer, J.-P.; Carrere, H. Effect of lignin-derived and furan compounds found in lignocellulosic hydrolysates on biomethane production. Bioresour Technol. 2012, 104, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Palmqvist, E.; Hahn-Hägerdal, B. Fermentation of lignocellulosic hydrolysates. II: Inhibitors and mechanisms of inhibition. Bioresour Technol. 2000, 74, 25–33. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, Z.; Wang, X.; Wang, N.; Wang, W.; Bao, J. Biodetoxification of toxins generated from lignocellulose pretreatment using a newly isolated fungus, Amorphotheca resinae ZN1, and the consequent ethanol fermentation. Biotechnol. Biofuels 2010, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Ran, H.; Zhang, J.; Gao, Q.; Lin, Z.; Bao, J. Analysis of biodegradation performance of furfural and 5-hydroxymethylfurfural by Amorphotheca resinae ZN1. Biotechnol. Biofuels 2014, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, M.T.; Ann Franden, M.; Johnson, C.W.; Beckham, G.T. Conversion and assimilation of furfural and 5-(hydroxymethyl)furfural by Pseudomonas putida KT2440. Metab. Eng. Commun. 2017, 4, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Koopman, F.; Wierckx, N.; De Winde, J.H.; Ruijssenaars, H.J. Identification and characterization of the furfural and 5-(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. Proc. Natl. Acad. Sci. USA 2010, 107, 4919–4924. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, B.; Ezeji, T.C. Biotransformation of furfural and 5-hydroxymethyl furfural (HMF) by Clostridium acetobutylicum ATCC 824 during butanol fermentation. New Biotechnol. 2012, 29, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Koopman, F.; Wierckx, N.; de Winde, J.H.; Ruijssenaars, H.J. Efficient whole-cell biotransformation of 5-(hydroxymethyl)furfural into FDCA, 2,5-furandicarboxylic acid. Bioresour. Technol. 2010, 101, 6291–6296. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Slininger, P.J.; Dien, B.S.; Berhow, M.A.; Kurtzman, C.P.; Gorsich, S.W. Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J. Ind. Microbiol. Biotechnol. 2004, 31, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Taherzadeh, M.J.; Gustafsson, L.; Niklasson, C.; Liden, G. Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2000, 53, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Bao, J.; Lu, J.; Lv, Q. Biodegradation of furfural by Bacillus subtilis strain DS3. J. Environ. Biol. 2015, 36, 727–732. [Google Scholar] [PubMed]
- Wierckx, N.; Koopman, F.; Ruijssenaars, H.J.; De Winde, J.H. Microbial degradation of furanic compounds: Biochemistry, genetics, and impact. Appl. Microbiol. Biotechnol. 2011, 92, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Nakatsukasa, W.M.; Mabe, J.A. Galactose induced colonial dissociation in Streptomyces aureofaciens. J. Antibiot. 1978, 31, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Lalucat, J.; Bennasar, A.; Bosch, R.; Garcia-Valdes, E.; Palleroni, N.J. Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 2006, 70, 510–547. [Google Scholar] [CrossRef] [PubMed]

 ) or furfural (●) as the sole carbon source. Cells were inoculated in M9 media at pH 7.5 containing NH4Cl (5 mM) as the nitrogen source. At the indicated times, the increments of cell growth of each culture were measured. A no inoculated flask containing culture medium was used as a control (result not shown). The experiment was carried out in triplicate obtaining similar results.
) or furfural (●) as the sole carbon source. Cells were inoculated in M9 media at pH 7.5 containing NH4Cl (5 mM) as the nitrogen source. At the indicated times, the increments of cell growth of each culture were measured. A no inoculated flask containing culture medium was used as a control (result not shown). The experiment was carried out in triplicate obtaining similar results.
 ) or furfural (●) as the sole carbon source. Cells were inoculated in M9 media at pH 7.5 containing NH4Cl (5 mM) as the nitrogen source. At the indicated times, the increments of cell growth of each culture were measured. A no inoculated flask containing culture medium was used as a control (result not shown). The experiment was carried out in triplicate obtaining similar results.
) or furfural (●) as the sole carbon source. Cells were inoculated in M9 media at pH 7.5 containing NH4Cl (5 mM) as the nitrogen source. At the indicated times, the increments of cell growth of each culture were measured. A no inoculated flask containing culture medium was used as a control (result not shown). The experiment was carried out in triplicate obtaining similar results.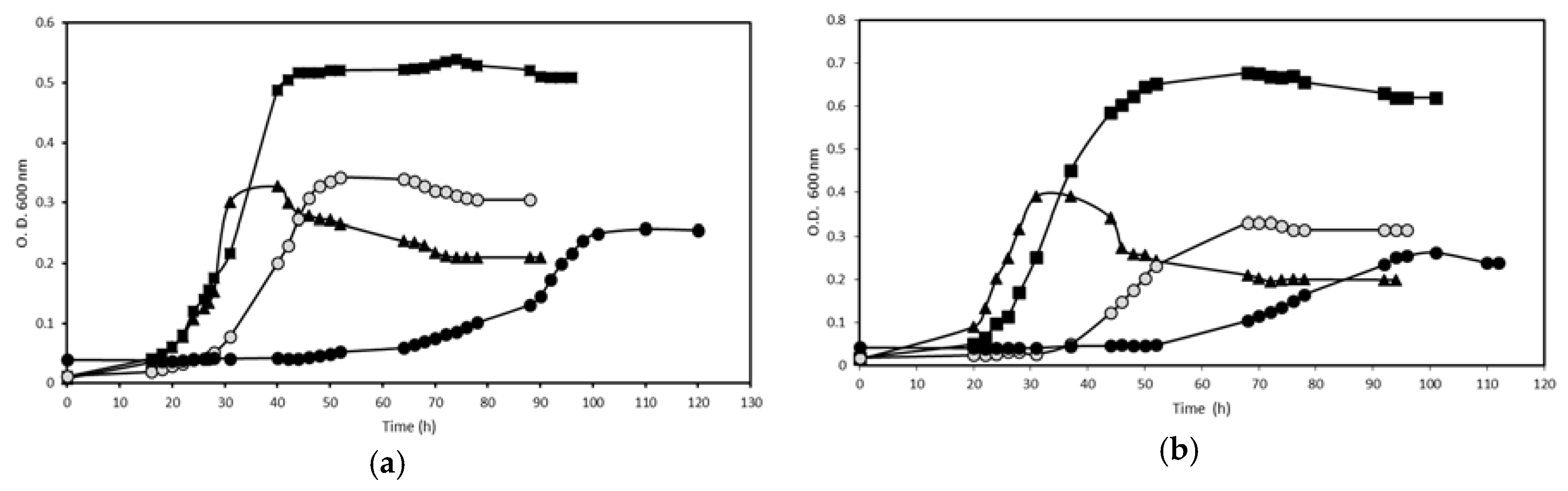
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Igeño, M.I.; Sánchez-Clemente, R.; Población, A.G.; Guijo, M.I.; Merchán, F.; Blasco, R. Biodegradation of 5-(Hydroxymethyl)-furfural and Furan Derivatives. Proceedings 2018, 2, 1283. https://doi.org/10.3390/proceedings2201283
Igeño MI, Sánchez-Clemente R, Población AG, Guijo MI, Merchán F, Blasco R. Biodegradation of 5-(Hydroxymethyl)-furfural and Furan Derivatives. Proceedings. 2018; 2(20):1283. https://doi.org/10.3390/proceedings2201283
Chicago/Turabian StyleIgeño, María Isabel, Rubén Sánchez-Clemente, Ana G. Población, M. Isabel Guijo, Faustino Merchán, and Rafael Blasco. 2018. "Biodegradation of 5-(Hydroxymethyl)-furfural and Furan Derivatives" Proceedings 2, no. 20: 1283. https://doi.org/10.3390/proceedings2201283
APA StyleIgeño, M. I., Sánchez-Clemente, R., Población, A. G., Guijo, M. I., Merchán, F., & Blasco, R. (2018). Biodegradation of 5-(Hydroxymethyl)-furfural and Furan Derivatives. Proceedings, 2(20), 1283. https://doi.org/10.3390/proceedings2201283





