Abstract
Cell registration by artificial neural networks (ANNs) in combination with principal component analysis (PCA) has been demonstrated for cell images acquired by light emitting diode (LED)-based compact holographic microscopy. In this approach, principal component analysis was used to find the feature values from cells and background, which would be subsequently employed as neural inputs into the artificial neural networks. Image datasets were acquired from multiple cell cultures using a lensless microscope, where the reference data was generated by a manually analyzed recording. To evaluate the developed automatic cell counter, the trained system was assessed on different data sets to detect immortalized mouse astrocytes, exhibiting a detection accuracy of ~81% compared with manual analysis. The results show that the feature values from principal component analysis and feature learning by artificial neural networks are able to provide an automatic approach on the cell detection and registration in lensless holographic imaging.
1. Introduction
In the last decade, automatic cell counting has become one of the most challenging task in the biomedical fields. Highly precise object recognition is a prerequisite for reliable cell monitoring. In this case, in terms of a continuous measurement, sophisticated automated image analysis methods have already been developed and used in several research laboratories. However, the cell images were captured using conventional microscopes, which are bulky, heavy, and expensive. The density, proliferation, and movement of the cells in such cultures should be closely investigated [1]. Therefore, using a lensless holographic microscope, which is compact and low-cost, the imaging at micro-scale can be provided continuously inside a cell incubator without a need for dismounting the monitored samples during characterization.
Inspired by human nervous system, the artificial neural network (ANN) methodology is a powerful tool to handle this kind of difficult and challenging problems and has been widely used to investigate the mechanism behind climate changes and even predict the climate change trend [2]. In this work, lensless holographic microscopy (Figure 1a) is combined with an automated image processing based on neural networks to offer a suitable solution fulfilling an automatic cell counting during cell culture. Since microscopic images provide high dimensional information vectors, we propose the use of principal component analysis (PCA) for the extraction of the feature values from the images of cells and non-cells (background), in order to reduce the complexity of the detection. The analysis and classification of the feature values are then performed by an ANN as a learning system, which is trained beforehand by a set of sample data. The neural network approach has been proven to be a fast, lightweight, and straightforward technique, which can be established with training and reference data instead of carefully crafted computational filters [3].
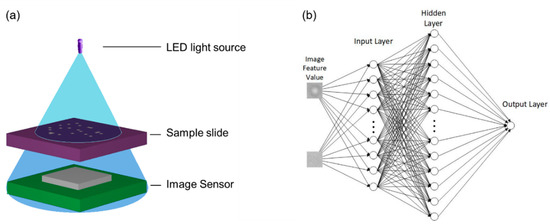
Figure 1.
(a) Schematic of the LED-based lensless holographic microscope setup. (b) The ANNs architecture.
2. Cell Counting Method
Digital images can be defined as a function of two variables, f(x, y), where x and y are spatial coordinates and the value of f(x, y) is the intensity of the image in the coordinates. In this work, readings from a digital-holographic microscope (DHM) in form of a monochrome microscope image are utilized. As a pre-processing step, two different input clusters of images are prepared as targets, both for positive cell detection and background images. The size of each input image is 50 × 50 pixels.
In order to reduce the input vector for the ANN-based classifier, PCA is used to analyze similarities and differences of the data. PCA is a popular and powerful tool for the analysis of high dimensional data with multiple applications to feature extraction and pattern detection [4]. In our approach, PCA is used to identify patterns on the image and represent the data into a vector based on important information. This can significantly minimize the size and complexity of the neural networks employed in cell classification problems, as well as reduce the training time and improve its overall performance significantly.
Given data vector x1 … xn from the input image, the sample mean (µz) is calculated by . As a result, the PCA is processed from 50 × 50 pixels to 50 × 1 vector. It is obvious that if we use and compute large number of vectors, computational complexity is increasing in the current and subsequent steps. Equation (1) is employed to calculate the sample covariant matrix,
where Cz and n are the covariant matrix and the number of the sample, respectively.
By projecting every input image on these cell images, 50 × 1 vectors will be directed to the ANN learning method. Algorithm based on Equation (2) is used to calculate updated weights,
where α, w, t and x are the learning rate, the weight, the target and the input, respectively.
Three layers of ANN will be trained using training cells and background images to minimize the error vector on its output layer. Each element of the output is a number between zero and one, which represents the similarity of input images to one of two classes (i.e., cell and non-cell). Iterative training of the neural networks with cell images will subsequently correct the weight values towards the desired target. The input images of cells and non-cells will be classified into classes, which have the greatest similarity to them. Figure 1b shows the architecture process classification using three layer neural networks.
3. Experimental Details
The experimental setup for the cell imaging by digital holography is depicted in Figure 1a showing the DHM as well as position of the cell sample. The DHM was utilized in this work to acquire the cell images that were used as both training data and ANN inputs. It consists mainly of an LED light source and a complementary metal oxide semiconductor (CMOS) image sensor [5]. In the DHM, different cell cultures including immortalized mouse astrocyte cultures were placed closely above the sensor and illuminated by the partially coherent LED light source. The diffraction pattern of the sample was recorded by a CMOS sensor and reconstructed by computation using the angular spectrum approach [6].
As input data preparation, we manually cut the input images to 50 × 50 pixel images of cells and non-cells (see Figure 2a–f) in order to obtain information details of the region of interest, or in other words, to eliminate the unnecessary data for the recognition. Afterwards, the image feature extractions from the microscopic data were performed by the PCA. In this experiment, we used both cell and background images as training data for the neural networks.
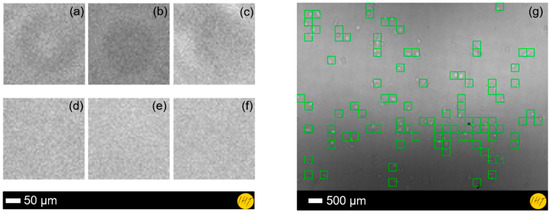
Figure 2.
PCA and ANN references for learning process: (a–c) 50 × 50 pixels cell images and (d–f) 50 × 50 pixels background images. (g) Result of the proposed ANN for the microscopic images showing its ability to detect 93 cells from a total number of 115 immortalized mouse astrocytes.
Moreover, the PCA reference was utilized to train the neuron image and update the weight value in ANNs. Our ANN has an input layer with 50 inputs, 400 neurons in a hidden layer, and 1 neuron in an output layer. For the activation function in ANNs, a sigmoid function was selected with one hidden layer and 105 iterations, respectively, for a maximum weight calculation iteration.
4. Results
The result reveals that our system is able to perform automatic cell counting by combining PCA and ANN techniques. In the learning process, the ANNs yielded a maximum error rate of 8 × 104. We have compared the computational performances of these algorithms using ANNs and manual calculation from the real data. From the system evaluation using different test images, a cell detection accuracy of ~81% could be achieved (see Figure 2g). The influence of the PCA was also investigated by comparing the results with a PCA-free classification, which could only detect cells with an accuracy of ~20%. The PCA features also show the ability of minimizing the processing time down to 13.84 s, compared to 18,960.03 s without PCA. Regardless of the required system improvement, for further studies and more accurate classification, the developed method has shown the feasibility to realize accurate automated cell detection and quantification for life science applications.
5. Conclusions
Artificial neural networks (ANNs) in combination with principal component analysis (PCA) have been employed for cell registration of cell images acquired by an LED-based compact lensless microscope. To evaluate the developed automatic cell counter, the trained system has been assessed on different datasets to detect immortalized mouse astrocytes cells. In this approach, PCA has been used to find the feature values from cells and background, which would be subsequently employed as neural inputs into the ANNs. Image datasets have been acquired from multiple cell cultures using a lensless microscope and compared with the reference data that have been generated by a manually analyzed recording. In this case, the experimental results have exhibited a detection accuracy of >80% compared with manual analysis, demonstrating the effectiveness of the proposed method for automatic cell counting in lensless imaging. Further enhancements are currently being performed to increase the accuracy and usability of the methods.
Author Contributions
Conceptualization, A.B.D., G.S., S.M. and H.S.W.; Methodology, A.B.D.; Software, A.B.D. and G.S.; Validation, J.H., P.H. and K.H.; Formal Analysis, G.S., I.A. and S.W.; Investigation, G.S. and H.S.W.; Resources, G.S.; Data Curation, A.B.D. and G.S.; Writing—Original Draft Preparation, A.B.D., G.S. and S.M.; Writing—Review and Editing, H.S.W., I.A., S.W. and J.D.P.; Visualization, A.B.D.; Supervision, H.S.W. and A.W.; Project Administration, H.S.W. and A.W.; Funding Acquisition, J.D.P., A.W. and H.S.W.
Funding
This work has been partially performed within LENA-OptoSense and QUANOMET funded by the Lower Saxony Ministry for Science and Culture (N-MWK) and European project of ChipScope funded by the European Union’s Horizon 2020 research and innovation program under grant agreement No. 737089.
Acknowledgments
The authors thank K.-H. Lachmund for the technical support. A.B.D thanks Kemenristekdikti-LPDP for the scholarship. S.M. thanks Georg-Christoph-Lichtenberg for the scholarship (Tailored Light).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wu, Y.; Ozcan, A. Lensless digital holographic microscopy and its applications in biomedicine and environmental monitoring. Methods 2018, 136, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Artificial Neural Network. In Multivariate Time Series Analysis in Climate and Environmental Research; Springer: Cham, Switzerland, 2018; ISBN 978-3-319-67340-0. [Google Scholar]
- Rivenson, Y.; Zhang, Y.; Günaydın, H.; Teng, D.; Ozcan, A. Phase recovery and holographic image reconstruction using deep learning in neural networks. Light Sci. Appl. 2018, 7, 17141. [Google Scholar] [CrossRef] [PubMed]
- Cardot, H.; Degras, D. Online Principal Component Analysis in High Dimension: Which Algorithm to Choose? Int. Stat. Rev. 2017, 86, 29–50. [Google Scholar] [CrossRef]
- Scholz, G.; Xu, Q.; Schulze, T.; Boht, H.; Mattern, K.; Hartmann, J.; Dietzel, A.; Scherneck, S.; Rustenbeck, I.; Prades, J.D.; et al. LED-based tomographic imaging for live-cell monitoring of pancreatic islets in microfluidic channels. Proceedings 2017, 1, 552. [Google Scholar] [CrossRef]
- Poon, T.C.; Liu, J.P. Introduction to Modern Digital Holography; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).