Regenerable Bead-Based Microfluidic Device with integrated THIN-Film Photodiodes for Real Time Monitoring of DNA Detection †
Abstract
:1. Introduction
2. Materials and Methods
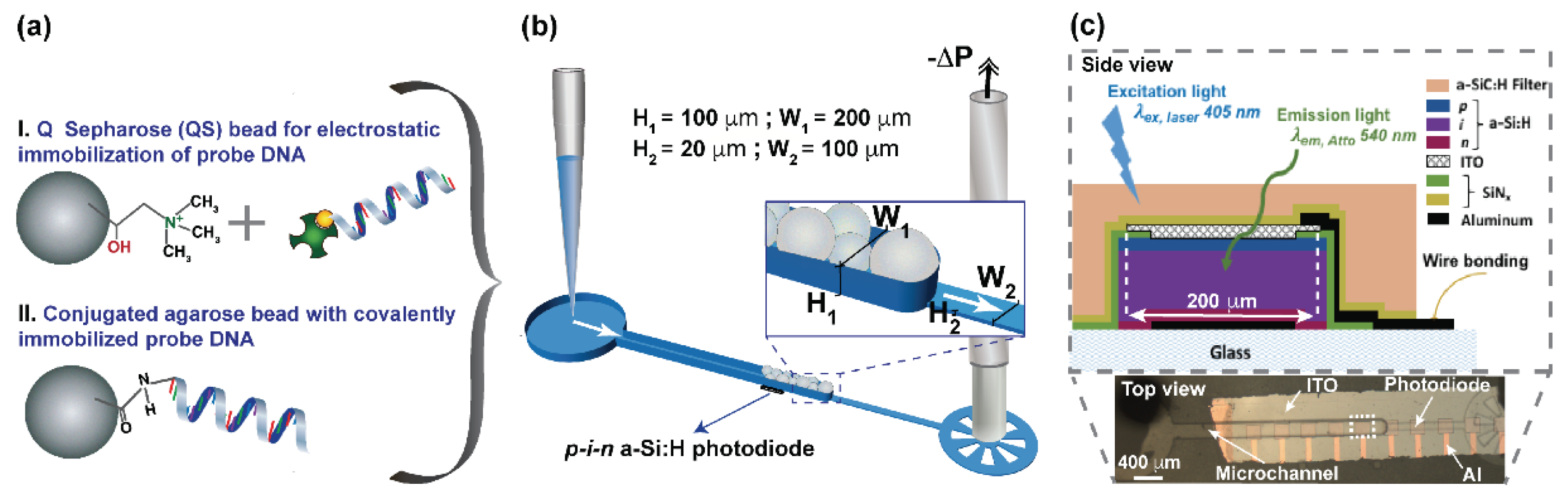
2.1. DNA Immobilization Strategies
2.2. Fabrication of Microchannel and Bead Trapping
2.3. Fabrication of a-Si:H Photodiodes
3. Results and Discussion
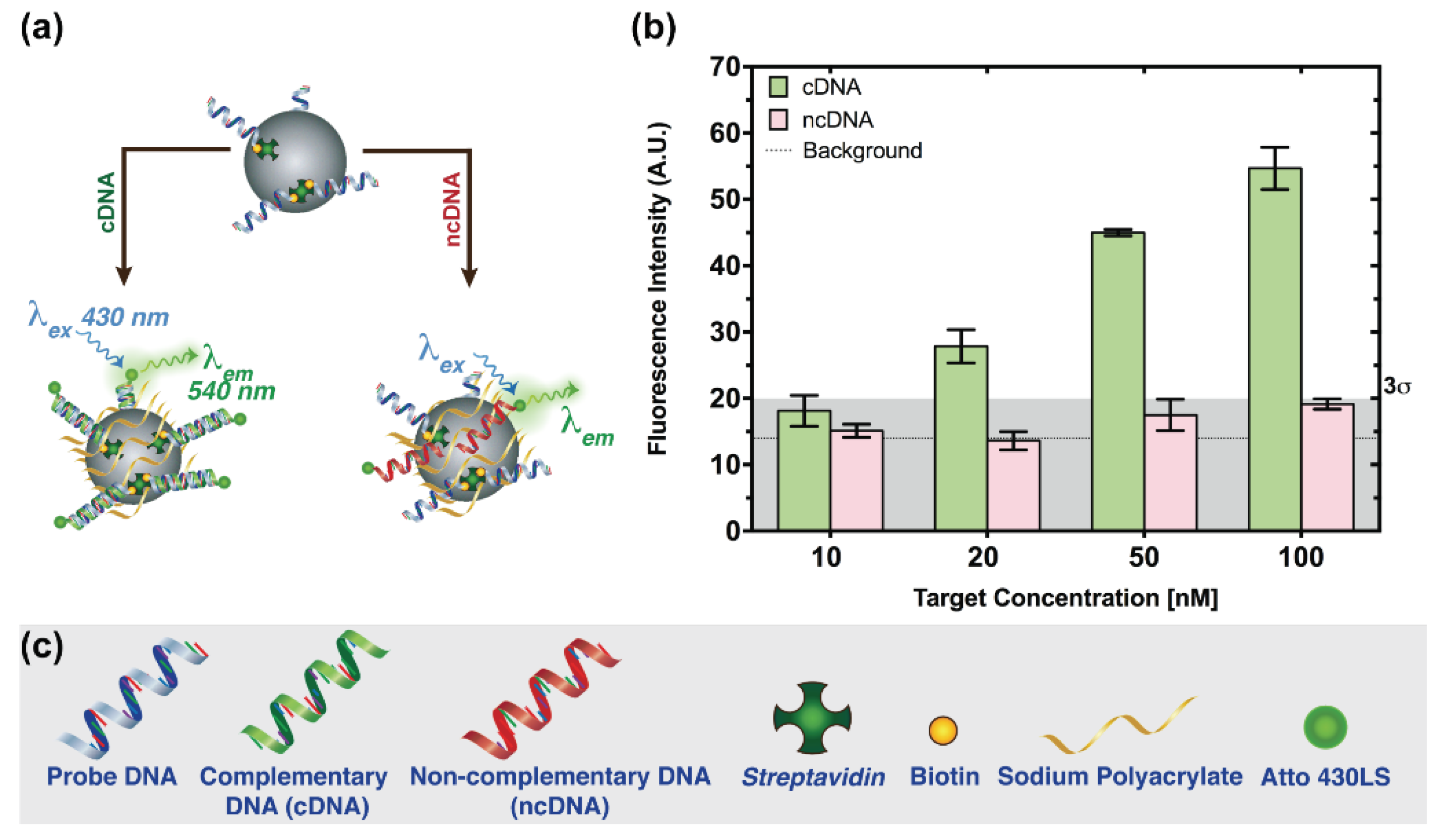
3.1. Probe Immobilization Strategies
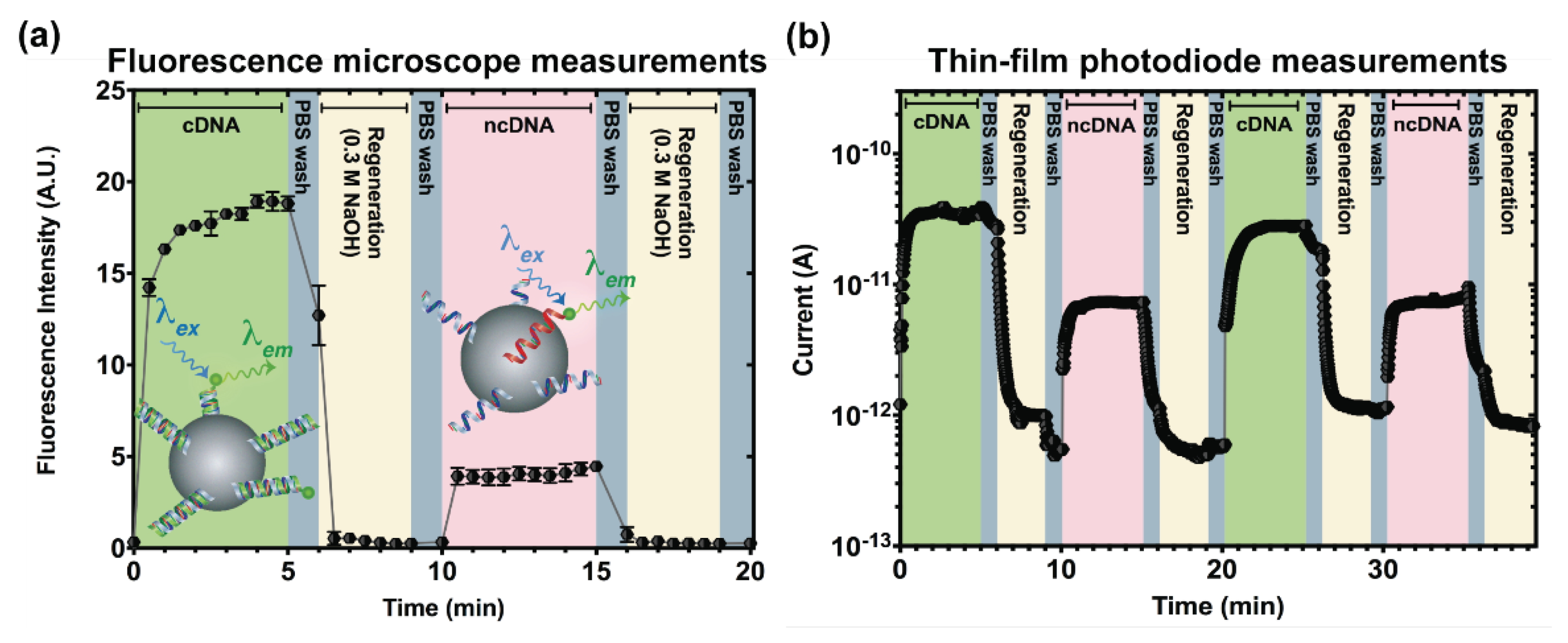
3.2. Microcolumn Regeneration
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ferguson, B.S.; Buchsbaum, S.F.; Swensen, J.S.; Hsieh, K.; Lou, X.; Soh, H.T. Integrated microfluidic electrochemical DNA sensor. Anal. Chem. 2009, 81, 6503–6508. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, Y.; Gan, W.; Liu, Y.; Xiang, G.; Wang, D.; Wang, L.; Cheng, J.; Liu, P. An automated microfluidic system for single-stranded DNA preparation and magnetic bead-based microarray analysis. Biomicrofluidics 2015, 9, 024102. [Google Scholar] [CrossRef] [PubMed]
- Pinto, I.F.; Caneira, C.R.; Soares, R.R.; Madaboosi, N.; Aires-Barros, M.R.; Conde, J.P.; Azevedo, A.M.; Chu, V. The application of microbeads to microfluidic systems for enhanced detection and purification of biomolecules. Methods 2017, 116, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Pinto, I.F.; Santos, D.R.; Caneira, C.R.F.; Soares, R.R.G.; Azevedo, A.; Chu, V.; Conde, J.P. Optical biosensing in microfluidics using nanoporous microbeads and amorphous silicon thin-film photodiodes: Quantitative analysis of molecular recognition and signal transduction. J. Micromech. Microeng. 2018, 28, 094004. [Google Scholar] [CrossRef]
- Santos, D.R.; Soares, R.R.G.; Chu, V.; Conde, J.P. Performance of hydrogenated amorphous silicon thin film photosensors at ultra-low light levels: Towards attomole sensitivities in lab-on-chip biosensing applications. IEEE Sens. J. 2017, 17, 6895–6903. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caneira, C.R.F.; Santos, D.R.; Chu, V.; Conde, J.P. Regenerable Bead-Based Microfluidic Device with integrated THIN-Film Photodiodes for Real Time Monitoring of DNA Detection. Proceedings 2018, 2, 953. https://doi.org/10.3390/proceedings2130953
Caneira CRF, Santos DR, Chu V, Conde JP. Regenerable Bead-Based Microfluidic Device with integrated THIN-Film Photodiodes for Real Time Monitoring of DNA Detection. Proceedings. 2018; 2(13):953. https://doi.org/10.3390/proceedings2130953
Chicago/Turabian StyleCaneira, Catarina R. F., Denis R. Santos, Virginia Chu, and João P. Conde. 2018. "Regenerable Bead-Based Microfluidic Device with integrated THIN-Film Photodiodes for Real Time Monitoring of DNA Detection" Proceedings 2, no. 13: 953. https://doi.org/10.3390/proceedings2130953
APA StyleCaneira, C. R. F., Santos, D. R., Chu, V., & Conde, J. P. (2018). Regenerable Bead-Based Microfluidic Device with integrated THIN-Film Photodiodes for Real Time Monitoring of DNA Detection. Proceedings, 2(13), 953. https://doi.org/10.3390/proceedings2130953




