Abstract
Nitrogen assimilation is vital for apple growth, yield, and quality, with nitrate reductase (NIA), nitrite reductase (NIR), glutamine synthetase (GS), and glutamate synthase (GOGAT) serving as key regulatory enzymes. This study systematically identified these four gene families in apple (Malus domestica) through genome-wide analysis and examined their expression patterns under nitrate treatment. In total, 13 genes were identified, 2 MdNIAs, 1 MdNIR, 7 MdGSs, and 3 MdGOGATs, with gene lengths ranging from 2577 to 27736 base pairs (bp); MdGLT1A had the longest coding sequence (6627 bp). The encoded proteins contained 355–2208 amino acids, with predicted isoelectric points (pIs) between 5.55 and 6.63. Subcellular localization analysis predicted distinct compartmentalization: MdNIA1A in peroxisomes; MdGS1 in the cytosol; MdNIR1, MdGS2, and MdGLU1 in chloroplasts; and MdGLT1 in mitochondria/chloroplasts. Functional site prediction revealed multiple phosphorylation and glycosylation sites, with ATP/GTP-binding motifs present only in certain MdGOGAT proteins. Protein interaction analysis suggested close associations among these genes and possible interactions with NRT2.1/2.2. Chromosomal mapping showed their distribution across eight chromosomes, while promoter analysis identified diverse cis-acting regulatory elements (e.g., ABRE and G-box). Under nitrate treatment (0–12 h), these genes exhibited distinct expression dynamics: MdNIA1A and B were rapidly induced (0–6 h) and maintained high expression; MdNIR1 peaked at 6 h and then declined; MdGS1.1B was activated after 6 h; and MdGS2A, MdGLU1, and MdGLT1A/B peaked at 6 h before decreasing. Therefore, these results elucidate the structural and functional divergence of nitrogen assimilation genes in apple and provide a basis for understanding nitrogen utilization mechanisms and developing nitrogen-efficient breeding strategies.
1. Introduction
Nitrogen is an essential nutrient for apple tree growth and development, and its efficient assimilation and utilization are key determinants of tree vigor, fruit yield, and quality and the ecological sustainability of orchards. However, excessive nitrogen fertilizer application in orchard systems leads to low nitrogen use efficiency, higher production costs, reduced fruit quality, and environmental pollution [1]. A comprehensive analysis of nitrogen assimilation in apples is, therefore, critical, given its complex molecular regulation involving several tightly coordinated enzymes. The enzymes nitrate reductase (NR), nitrite reductase (NiR), glutamine synthase (GS), and glutamate synthase (GOGAT) form a four-step, rate-limiting reaction sequence central to the plant nitrogen assimilation pathway. In most terrestrial plants, including apple trees, nitrate serves as the primary nitrogen source [2]. NR catalyzes the reduction of nitrate (NO3−) to nitrite (NO2−), which is then reduced to ammonium (NH4+) by NiR in plastids. NH4+ is incorporated into glutamine (Gln) and glutamate (Glu) via the GS/GOGAT cycle—core reactions that provide nitrogen skeletons for synthesizing amino acids, nucleic acids, and secondary metabolites [3]. Therefore, elucidating the structure, function, gene regulation, and environmental interactions of key enzymes involved in apple nitrogen assimilation is crucial. Such studies will deepen our understanding of nitrogen metabolism in apples, support the development of precise fertilization strategies, enable the breeding of nitrogen-efficient cultivars, and promote the sustainable advancement of the apple industry.
Cytoplasmic nitrate reductase (NR) is a rate-limiting enzyme in plant nitrogen metabolism, directly regulating this process [4]. NR is a dimeric protein complex, with each identical subunit containing three cofactors: a molybdenum cofactor (MoCo), a heme-binding domain (Heme), and a flavin adenine dinucleotide-binding domain (FAD) [5]. In Arabidopsis, two NR isoforms—NIA1 and NIA2—are encoded by the AtNIA1 and AtNIA2 genes, respectively [6,7]. Among them, NIA2 plays a critical role in nitrogen assimilation and stress tolerance [8]. The nia1/nia2 double mutant exhibits severe growth retardation and nitrate accumulation, highlighting the enzyme’s essential function [9]. In rice, three genes encode NR, with OsNIA1 contributing predominantly to overall enzyme activity [10]. Functional analysis of OsNR2 has shown that it enhances effective tiller number, grain yield, and nitrogen fertilizer use efficiency in japonica rice [11]. In apples, the MdNIA2 gene improves salt and drought tolerance in Arabidopsis and callus cultures by modulating nitrogen use efficiency and nitric oxide (NO) metabolism.
The plastid nitrite reductase (NiR) converts NO2− produced by NR into NH4+ and functions within plastids. NIR1 is a key regulator of nitrogen assimilation, NO homeostasis, and plant growth stress responses [12]. The rice OsNiR promoter contains a nitrate-responsive cis-element (NRE). Overexpression of OsNiR increases tiller number and overall yield, thereby enhancing nitrogen use efficiency under both high- and low-nitrogen conditions [13]. Research on NiR in fruit trees remains limited, with PtNiR identified only once—in Citrus under drought stress [14].
Glutamine synthetase (GS) catalyzes the assimilation of NH4+ to produce glutamine. In plants, two functional GS types exist: the prokaryotic GSIIb (GLN2) and the eukaryotic GSIIe (GLN1/GS). Angiosperms generally possess two nuclear gene classes encoding glutamine synthetase II enzymes (GSIIes): cytoplasmic glutamine synthetase (GS1) and plastidic glutamine synthetase (GS2) [15,16]. GS2 is predominantly localized in photosynthetic tissues such as mesophyll cells [1,17,18], whereas GS1 is ubiquitously present across various plant organs and tissues [3]. Simultaneous overexpression of OsGS1;1 and OsGS2 in rice improves nitrogen use efficiency (NUE) [19]. In maize, two cytoplasmic glutamine synthetases, GS1-3 and GS1-4, have been identified. Knockout studies show that these enzymes have distinct, non-redundant roles in determining grain yield: GS1-3 primarily affects grain number, while GS1-4 influences grain size [20]. In the apple rootstock, genes associated with nitrogen metabolism, such as MdGS1, are downregulated [21].
Glutamate synthase (GOGAT) catalyzes the synthesis of glutamic acid from glutamine and α-ketoglutarate and exists in two forms: ferredoxin-dependent (Fd-GOGAT) and NADH-dependent (NADH-GOGAT) [22]. NADH-GOGAT is predominantly found in non-photosynthetic tissues [23,24]. In Arabidopsis, two genes, Glu1 and Glu2, encode Fd-GOGAT, while in rice, OsFd-GOGAT mutants exhibit chlorosis under natural conditions [25]. Conversely, overexpressing NADH-GOGAT in rice enhances grain weight and improves nitrogen utilization [26]. In apple, studies mainly examine how trace elements regulate GOGAT activity during nitrogen metabolism. Under aluminum stress, GOGAT activity in roots and leaves is significantly inhibited. However, magnesium supplementation restores this activity, promoting glutamate synthesis and improving nitrogen metabolism [27]. Similarly, calcium treatment mitigates temperature-induced stress on nitrogen metabolism in apple roots by regulating GOGAT activity [28].
Despite extensive research on nitrogen assimilation in model plants such as Arabidopsis and rice, these processes remain poorly understood in perennial fruit trees like apple. Apple is an economically important fruit crop worldwide with significant nutritional and commercial value. Identifying and characterizing its nitrogen-assimilation-related genes are crucial for elucidating nitrogen utilization efficiency mechanisms and developing strategies for improving apple production and sustainability. This study aimed to systematically identify and characterize the four key nitrogen assimilation enzyme gene families (NIA, NIR, GS, and GOGAT) in apple (Malus domestica) through genome-wide analysis. We explored their structural features, subcellular localization, protein interaction networks, promoter cis-acting elements, and expression profiles under nitrate treatment. These findings provide a foundation for understanding nitrogen utilization in apple and for guiding the development of nitrogen-efficient breeding strategies.
2. Materials and Methods
2.1. Identification of Apple Nitrogen Key Enzyme Genes
The genome of the apple (Malus domestica) used in this study is the ‘Golden Delicious’ apple (GDDH13) reference genome (GDDH13 v1.1, https://iris.angers.inra.fr/gddh13/, accessed on 26 January 2025) [29]. Two query strategies (blastp and hmmsearch) are used to identify members in this article. The query sequence used for protein homologous alignment is the protein sequence of the pattern species Arabidopsis thaliana (L.) Heynh. The sequence is downloaded from the TAIR database (https://www.arabidopsis.org/, accessed on 26 January 2025). After screening, the searched apple members were submitted to the SMART website (http://smart.embl-heidelberg.de/, accessed on 26 January 2025) for conservative domain confirmation, and we finally obtained the candidate gene [30].
2.2. Characteristic Analysis of Protein
To calculate the molecular weight and theoretical isoelectric point (pI), bioinformatic analysis was carried out with the ExPASy online tool (https://web.expasy.org/protparam/, accessed on 31 January 2025).
2.3. Subcellular Localization and Signal Peptide Prediction of Protein
The online program WoLFPSORT: Protein Subcellular Localization Prediction (https://wolfpsort.hgc.jp/, accessed on 31 January 2025) was used to predict protein subcellular localization, and the online website SignalP (https://services.healthtech.dtu.dk/services/SignalP-4.1/, accessed on 31 January 2025) was used to identify lipoprotein signal peptides [31].
2.4. Active Site Prediction of Protein
The PROSITE Server (http://www.expasy.org/prosite/, accessed on 14 March 2025) was used to find protein functional sites (glycosylation sites, acylation sites, phosphorylation sites, etc.) [32]. The NetPhos 3.1 Server (https://services.healthtech.dtu.dk/services/NetPhos-3.1/, accessed on 14 March 2025) was used to predict and analyze the number of phosphorylation sites of three amino acids (serine, threonine, and tyrosine) [33].
2.5. Protein Interaction Network
Analysis of the interaction network was conducted through the STRING functional protein association network (https://string-db.org, accessed on 21 March 2025) and the species source selection model species Arabidopsis.
2.6. Gene Structure Analysis of Apple Nitrogen Key Enzyme Genes
Using the Gene Structure View package of TBtools (v1.098691), the CDS, UTR, and intron configurations of apple nitrogen key enzyme genes were examined with an apple genome annotation file (Malus domestica.v1.0.consensus.gff) [34].
2.7. Chromosome Localizations of Apple Nitrogen Key Enzyme Genes
The location information of apple nitrogen key enzyme genes was extracted from the gene ID in the apple genome annotation file (Malus domestica.v1.0.consensus.gff), and submitted to the online software MG2C (http://mg2c.iask.in/mg2c_v2.0/, accessed on 31 January 2025) for chromosome mapping.
2.8. Promoter Cis-Acting Element Prediction of Apple Nitrogen Key Enzyme Genes
Fragments about 2000 bp upstream of the translation start site of the apple nitrogen key enzyme genes from the apple genomic sequence (GDDH13_1-1_formatted.fasta) used as the promoter regions, and the gene promoter sequence identified above, were submitted to Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 6 February 2025) for predictive analysis of the promoter cis-acting elements.
2.9. Expression of Key Apple Nitrogen Enzyme Genes Under Different Nitrate Treatments (Including Nitrogen Starvation Treatment)
From the seedlings of ‘Malus hupehensis’, 18 seedlings with relatively consistent growth status were selected. The seedlings were first pre-cultured with nitrogen-deficient MS culture medium (ingredients: 1.2 mM KH2PO4, 0.1 mM CaCl2, 1 mM MgSO4, 0.1 mM Fe salt, trace amount, 1 mM KCl) for one week, and then, 5 mM KNO3 was added for nitrogen treatment. The entire plant was sampled for 0, 3, 6, and 12 h during the nitrogen treatment.
The total RNA was extracted using the TRIzol Reagent kit (TakaRa, Dalian, China), and the reverse transcription and synthesis of cDNA were performed using the Prime Script RT Reagent Kit with the gDNA Eraser kit. Real-time fluorescence quantitative PCR (qRT-PCR) was carried out according to the method provided by the Ultra SYBR Mixture (High ROX) Reagent (Vazyme Biotech, Nanjing, China). The reaction system was as follows: 2 × UltraSYBR Mixture (High ROX) 10 μL, ddH2O 7 μL, upstream and downstream primers (10 mM), and cDNA template at 1 μL; the total system was 20 μL. The qRT-PCR reaction process is as follows: pre-denaturation at 95 °C for 10 min, denaturation at 95 °C for 15 s, annealing at 58 °C for 15 s, and extension at 65 °C for 10 s; 40 cycles were performed. The fluorescence signal was collected in step 3 of each cycle, and each reaction was repeated 3 times. The design of quantitative primers was verified by the online website Primer3Plus (https://www.primer3plus.com/, accessed on 20 May 2025). All primer specificities were verified by Nucleotide BLAST and Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 20 May 2025) of the NCBI. The primers used for the reaction are displayed in Table S1; Md18S is the apple intrare gene. Data analysis of relative gene expression amounts was made at 2−ΔΔCT.
2.10. Statistical Analysis
Each experiment was independently performed in triplicate unless stated otherwise. The significant difference analysis and column chart creation were performed with DPS (DPS software, Sinyosoft, version 15.10, http://www.dpsw.cn/dps_eng/index.html, accessed on 10 June 2025) and GraphPad Prism 7 Software (GraphPad Software, La Jolla, CA, USA). Data were analyzed by one-way ANOVA with Tukey’s multiple comparison test, presented as means ± SD. Significant differences are indicated by different letters at the p < 0.05 level.
3. Results
3.1. Identification and Basic Information of Apple Nitrogen Assimilation Enzyme Genes
After strict screening and confirmation, a total of two MdNIA genes, one MdNIR gene, seven MdGS genes, and three MdGOGAT genes were obtained. Statistics showed that the length of each gene sequence is between 2577 and 27736 bp, and the length of the MdGLT1A (MD01G1219500) coding sequence of the MdGOGAT family reaches 6627 bp. The protein size is between 355 and 2208 aa, and the pI value is acidic in 5.55–6.63. See Table 1 for specific information.

Table 1.
Information about the NIA, NIR, GS, and GOGAT members found in apple.
3.2. Active Site Prediction of Apple Nitrogen Assimilation Enzymes
Protein functional sites include various types, such as glycosylation sites, phosphorylation sites, and myristoylation sites. Understanding these active sites can help further study and analyze protein function. In Table 2, we predict the protein active sites of apple nitrogen assimilation enzymes. There are many active sites of various types in MdNIA, and there are many protein kinase C phosphorylation sites in part of MdGOGAT members. The ATP/GTP-binding site motif A (P ring) is also only present in some MdGOGAT members.

Table 2.
Active site prediction of nitrogen assimilation enzymes in apple.
3.3. Prediction of Subcellular Localization and Signal Peptide Prediction of Apple Nitrogen Assimilation Enzyme Genes
Subcellular localization prediction can be used to infer the precise location of proteins in cells through calculation models. We found that MdNIA1A is mainly located in the peroxisome, MdNIA1B is mainly located in the cytoplasm, MdNIR1 is mainly located in the chloroplast, MdGS1 members are mainly located in the cytoplasm, MdGS2 is mainly located in the chloroplast (Table 3), MdGLU1 is mainly located in the chloroplast, and MdGLT1A and MdGLT1B are mainly located in the mitochondria and chloroplast. The signal peptide cleavage site of MdNIR1 may be between amino acids at positions 18 and 19 (Figure 1).

Table 3.
Subcellular localization and signal peptide of nitrogen assimilation enzymes in apple.
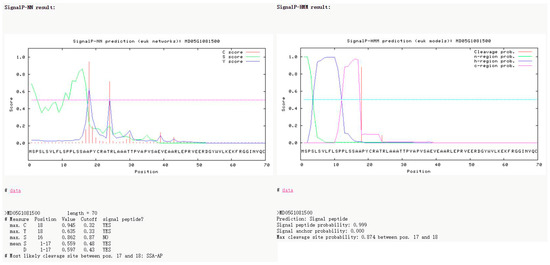
Figure 1.
MdNIR1 signal peptide prediction.
3.4. Construction of Interaction Protein Network for Nitrogen Assimilation Enzymes in Apple
In order to predict the potential biological function of apple nitrogen assimilation enzymes, a protein interaction relationship network of key enzymes of nitrogen assimilation in apples was constructed based on the homologous sequences in the model species Arabidopsis. In Figure 2, the results show that there is a very close relationship between the proteins of the key enzymes of nitrogen assimilation, and they all have a possible interaction with NRT2.1 and NRT2.2.
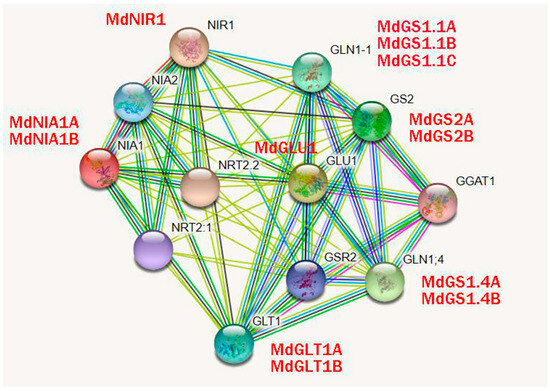
Figure 2.
Nitrogen assimilation enzyme protein interaction network in apples and Arabidopsis.
3.5. Chromosome Localization and Gene Structural Analysis of Apple Nitrogen Assimilation Enzyme Genes
Chromosome localization and gene structure analysis were performed on apple nitrogen assimilation enzyme genes using apple genome annotation (Malus domestica.v1.0.consensus.gff). Through comparative analysis, it was found that the distribution of each gene was relatively scattered. Among them, MdGLT1.1 is located in chromosome 1, MdNIR1 is located in chromosome 5, MdGLT1.2 is located in chromosome 7, MdNIA1.1 is located in chromosome 8, MdGS1.1A is located in chromosome 9, MdGS1.4A and MdGS2.1 are located in chromosome 13, MdGLU1 and MdGS1.1B are located in chromosome 14, MdNIA1.2 is located in chromosome 15, MdGS14B and MdGS2.2 are located in chromosome 16, and MdGS1.1C is located in chromosome 17 (Figure 3). The introns and exons of each gene were analyzed; the results showed that there were four exons in MdNIA1A, MdNIA1B, and MdNIR1, and the number of exons in the remaining genes was significantly larger (Figure 4).
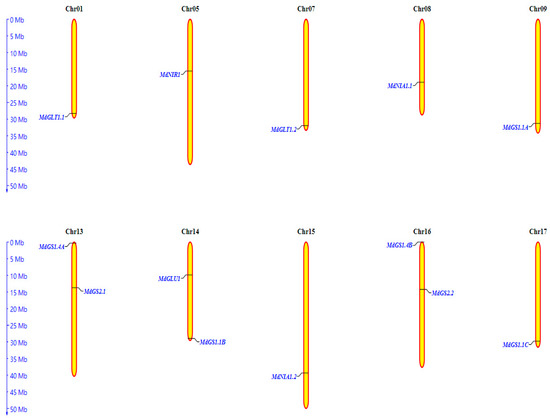
Figure 3.
Chromosomal location of apple nitrogen assimilation enzyme genes.
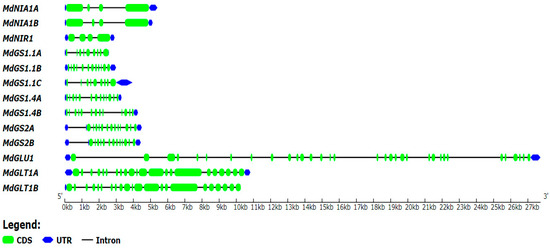
Figure 4.
Gene structure (intron/exon) of apple nitrogen assimilation enzyme genes.
3.6. Analysis of Cis-Acting Elements in the Promoter of Apple Nitrogen Assimilation Enzyme Genes
The specific types of cis-acting elements present in the promoter region of a gene play a crucial role in determining the regulatory factors that influence gene expression. To investigate the potential regulatory factors associated with the nitrogen assimilation enzyme genes in apple, we performed a predictive analysis of the cis-acting elements within their promoters. In Table 4, the analysis revealed that MdNIA1A contained a maximum of 5 ERF elements, MdNIA1B had up to 10 ABRE elements, and MdNIR1 possessed a maximum of 6 ERF elements. In MdGS1.1A, there were up to five ABRE elements, and MdGS1.1B contained three ABER, ARE, and G-box elements each, while MdGS1.1C exhibited a maximum of three ARE and CAT-box elements. MdGS1.4A had up to three ABRE, three CGTCA-motif, and three TGACG-motif elements, and MdGS1.4B had up to five ABRE elements. MdGS2A contained a maximum of three ABRE and W-box elements, and MdGS2B had up to three G-box elements. MdGLU1 exhibited a maximum of three CGTCA-motif and TGACG-motif elements. MdGLT1A contained up to 5 ABRE, CGTCA-motif, and TGACG-motif elements, and MdGLT1B had a maximum of 14 LTR elements. Taken together, the presence of these cis-acting elements suggests that the nitrogen assimilation enzyme genes may be involved in multiple responses, which require further studies to elucidate.

Table 4.
Cis-acting elements of apple nitrogen assimilation enzyme gene promoters. Promoter analysis was performed on 2000 bp sequences upstream of the transcription start sites.
3.7. Expression Analysis of Apple Nitrogen Assimilation Enzyme Genes Under Nitrate Nitrogen Treatment
Following the treatment of apple seedlings with nitrate nitrogen for varying durations (0, 3, 6 and 12h), the expression levels of 13 genes associated with apple nitrogen assimilation enzymes were analyzed, revealing diverse patterns of change (Figure 5). The expression profiles of the MdNIA1A and MdNIA1B genes were analogous, exhibiting a marked increase in expression from 0 to 6 h, followed by a relatively stable high expression from 6 to 12 h. This suggests that nitrate nitrogen treatment significantly induced transcription in the early phase (0–6h), with expression stabilizing in the later phase. The MdNiR1 gene expression was slightly upregulated between 0 and 6 h, decreased from 6 to 12 h, and peaked at 6 h, indicating that the gene’s expression activation in response to nitrate nitrogen was primarily concentrated around 6 h post-treatment, after which expression was inhibited. The gene expression of MdGS1.1A, MdGS1.1C, and MdGS1.4A demonstrated an overall upward trend, with rapid upregulation from 0 to 6 h and sustained high levels from 6 to 12 h, suggesting that nitrate treatment facilitated their continuous expression, with approximately 6 h identified as a critical induction point. The expression profile of the MdGS1.1B gene remained notably low and stable from 0 to 6 h, followed by a marked increase from 6 to 12 h. This suggests a delayed transcriptional response to nitrate nitrogen, with significant activation occurring after 6 h. Conversely, the MdGS1.4B gene exhibited a gradual increase in expression from 0 to 6 h, peaking at 6 h, and subsequently declining sharply by 12 h. This pattern is indicative of a typical short-term high induction followed by rapid attenuation. The expression levels of MdGS2A, MdGLU1, MdGLT1A, and MdGLT1B demonstrated an initial increase, peaking around 6 h, before declining, suggesting that these genes are upregulated by nitrate nitrogen between 0 and 6 h, with a subsequent downregulation from 6 to 12 h. This indicates that their response to nitrate nitrogen is most pronounced at the midpoint of the treatment (6 h), with negative regulation occurring thereafter. The expression of the MdGS2B gene showed a slight decrease between 0 and 3 h, an upregulation to a peak between 3 and 6 h, and a decline from 6 to 12 h. This gene’s response dynamics differ from the others, with the 3 to 6 h window being critical for activation. In summary, there are significant differences in the response patterns of different nitrogen assimilation-related enzyme genes to nitrate nitrogen treatment.
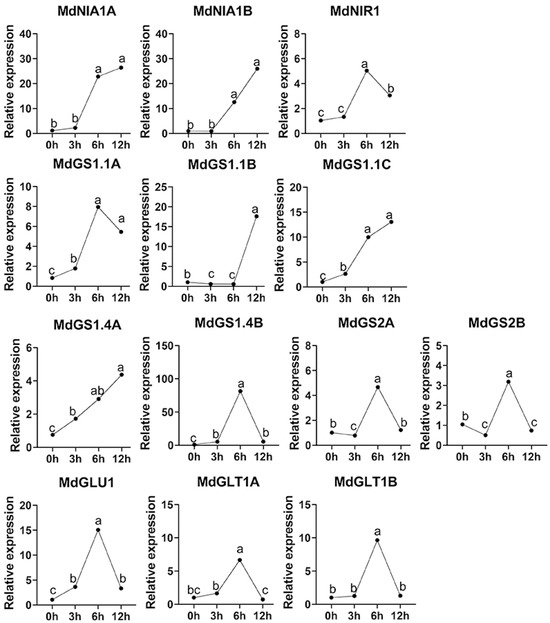
Figure 5.
Real-time fluorescence quantitative PCR primers for relative expression of apple nitrogen assimilation enzymes. Md18S was used as a control gene. One-way ANOVA with Tukey’s multiple comparison test shows significant differences at p < 0.05, marked by different letters, n ≥ 3.
4. Discussion
The present study provides a comprehensive genomic and expression analysis of four key nitrogen assimilation enzyme families (NIA, NIR, GS, and GOGAT) in apple (Malus domestica). By systematically characterizing 13 genes encoding these enzymes, we elucidated their structural diversity, subcellular localization, cis-regulatory elements, and dynamic expression patterns in response to nitrate treatment. This study addresses a significant knowledge gap in nitrogen metabolism in perennial fruit trees and offers new insights into the molecular mechanisms governing nitrogen use efficiency (NUE) in apple.
4.1. Gene Identification and Subcellular Localization Analysis
We identified 13 nitrogen-assimilation-related genes in apple—2 MdNIAs, 1 MdNIR, 7 MdGSs, and 3 MdGOGATs—highlighting the evolutionary expansion and functional diversification of these families in woody plants. Gene lengths varied from 2577 to 27,736 bp, and encoded proteins displayed distinct isoelectric points (pI 5.55–6.63), suggesting adaptive specialization and potential tissue- or context-specific regulatory roles (Table 1). For instance, the unusually long coding sequence of MdGLT1A (6627 bp) may reflect complex post-transcriptional regulation or protein interactions, warranting further functional studies (Table 1). Notably, plant NIR enzymes differ from microbial counterparts; for example, Bacillus tequilensis A2 NiR-A2 contains [Fe-S] cluster-binding domains and NADH-binding motifs, with optimal activity at 40 °C and pH 6.0 [35]. In contrast, our study revealed that MdNIR1 is chloroplast-localized with a pI of 6.63 (Table 1 and Table 3).
Subcellular localization predictions revealed patterns consistent with enzyme function. MdNIA1A localizes to peroxisomes and MdNIR1 to chloroplasts (Table 3), aligning with their sequential roles in nitrate reduction (NO3− → NO2− → NH4+). MdGLT1 exhibits dual localization in mitochondria and chloroplasts, suggesting a role in coordinating energy metabolism and nitrogen recycling (Table 3). Multiple MdGS isoforms show compartment-specific localization, MdGS1 in the cytosol and MdGS2 in chloroplasts (Table 3), indicating specialized ammonium assimilation pathways and enhancing nitrogen metabolism through compartmentalized carbon–nitrogen interactions.
The 13 genes are distributed across eight chromosomes, identifying potential targets for marker-assisted selection. For example, the co-localization of MdGS1.4A and MdGS2A on chromosome 13 could facilitate pyramiding favorable alleles to improve NUE (Figure 3). Future studies should assess allelic variation and haplotype diversity among apple cultivars.
4.2. Regulatory Networks and Expression Dynamics
Promoter analysis identified diverse cis-regulatory elements, including ABRE, ERE, and G-box motifs, indicating complex transcriptional regulation by hormones (ABA and ethylene), light, and stress signals. The high frequency of ABRE motifs in MdNIA1A/B and MdGLT1A/B suggests potential cross-talk between nitrogen assimilation and abiotic stress responses (Figure 4), consistent with findings that ABA modulates nitrate reductase activity in Arabidopsis [1]. Light-responsive elements, particularly the G-box in MdNIR1 and MdGS2, suggest coordination between nitrogen metabolism and photosynthesis (Figure 4), as observed in Arabidopsis, rice, and wheat [2].
The divergent expression patterns observed under nitrate treatment (0–12 h) indicate a temporally coordinated cascade of nitrogen assimilation. Early induction of MdNIA1A/B (0–6 h) promotes rapid nitrate reduction to nitrite, providing substrates for MdNIR1, which peaks at 6 h (Figure 5). This sequential activation mirrors the metabolic progression (NO3− → NO2− → NH4+) and highlights efficient substrate channeling. The delayed induction of MdGS1.1B (beyond 6 h) suggests a role in later stages of nitrogen remobilization or nitrogen reallocation under stress (Figure 5). Meanwhile, transient peaks of MdGS2A, MdGLU1, and MdGLT1A/B at 6 h indicate involvement in mid-stage ammonium assimilation, likely supporting rapid amino acid synthesis necessary for growth (Figure 5). These patterns contrast with rice, where OsGS1;1 and OsGS2 are co-induced, indicating distinct regulatory strategies in apple [19].
4.3. Gene Function Comparison and Interaction Networks
Comparative analysis of nitrogen assimilation between apple and model species such as Arabidopsis and rice reveals both conserved and divergent features. Notably, apple’s single MdNIR1 aligns with the monocot pattern exemplified by rice OsNIR (Table 1), whereas the presence of two MdNIA genes reflects the dicot-specific duplication seen in Arabidopsis (NIA1/NIA2) (Table 1) [8,13]. Additionally, the expansion of the MdGS family to seven members and the MdGOGAT family to three members exceeds Arabidopsis (GS: 5; GOGAT: 2 Fd-GOGAT, 1 NADH-GOGAT) (Table 1), suggesting adaptations to the metabolic demands of woody growth and fruit development [36,37]. The functional divergence and dynamic expression of these genes present promising targets for enhancing nitrogen use efficiency (NUE) in breeding programs. Constitutive overexpression of MdNIA1A/B, which both maintain high expression under nitrate treatment, could improve early nitrate reduction (Figure 5). In contrast, modulating MdGS1.1B or MdGS2A/B, which show delayed or transient induction, may optimize ammonium assimilation without imposing metabolic stress (Figure 5).
The predicted protein interaction network indicates links between nitrogen assimilation enzymes and nitrate transporters (NRT2.1/2.2), highlighting integrated regulation of nitrogen uptake and assimilation (Figure 2). This aligns with findings in Arabidopsis, where NRT2-mediated nitrate uptake is closely coordinated with nitrate reductase activity via feedback mechanisms [9]. Experimental validation of these interactions using co-immunoprecipitation or yeast two-hybrid assays could further elucidate the coordination of nitrogen transport and metabolism in apple.
This study identified key genes involved in nitrogen assimilation in apple. Analysis of their structural and functional diversity, along with examination of their expression patterns under nitrate treatment, provides a foundation for improving nitrogen use efficiency in apple cultivation. Future research can leverage these insights to reduce nitrogen fertilizer input without compromising yield or fruit quality, supporting sustainable agricultural practices.
5. Conclusions
This study systematically examined 13 nitrogen assimilation-related genes (2 MdNIAs, 1 MdNIR, 7 MdGSs, and 3 MdGOGATs) in apple, revealing structural variation and distinct expression patterns in response to nitrate treatment. Subcellular localization and promoter cis-element analyses further highlight their specialized functions. These genes represent potential targets for breeding nitrogen-efficient apple varieties. Future work should focus on functional validation, mapping regulatory networks, and conducting field trials to evaluate practical applications in sustainable apple production. Such advances could reduce fertilizer dependence while maintaining yield and quality, promoting eco-friendly orchard management.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/nitrogen6040099/s1: Table S1: Primers used in this study.
Author Contributions
Conceptualization, Y.L. and H.J.; methodology, T.L. and Z.F.; software, T.L., L.L. and Z.L.; validation, Q.Z., S.M. and X.L., H.G. and M.Z.; formal analysis, T.L., L.L. and Z.L.; investigation, T.L. and Z.F.; resources, Y.L. and H.J.; data curation, Y.Z. and S.W.; writing—original draft preparation, T.L.; writing—review and editing, T.L., Y.L. and H.J.; visualization, T.L. and Z.F.; supervision, Y.L. and H.J.; project administration, Y.L., H.J., Y.Z. and S.W.; funding acquisition, Y.L., H.J., Y.Z. and S.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of Shandong Province (2024CXGC010903 and 2023CXPT013), the National Key Research and Development Program (2023YFD2301000), the National Natural Science Foundation of China (32472705, 32402536, and 32302513), the Natural Science Foundation of Shandong Province (ZR2022JQ14 and ZR2022QC112), the National Industry Technology System of Apple (CARS-27), the Young Talent of Lifting Engineering for Science and Technology in Shandong (SDAST2024QTA083), and the Taishan Scholar Project.
Data Availability Statement
All data supporting the findings of this study are available in this paper and the Supplementary Information.
Acknowledgments
We sincerely thank our team leader Yuanyuan Li, for her great achievement and the support in our work. We also thank Han Jiang, Yali Zhang and Shang Wu for providing great funding supports.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Tegeder, M.; Masclaux-Daubresse, C. Source and Sink Mechanisms of Nitrogen Transport and Use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant Nitrogen Assimilation and Use Efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- Lea, P.J.; Miflin, B.J. Nitrogen Assimilation and Its Relevance to Crop Improvement. In Annual Plant Reviews, Volume 42: Nitrogen Metabolism in Plants in the Post-Genomic Era; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar]
- Aslam, M.; Huffaker, R.C. Role of Nitrate and Nitrite in the Induction of Nitrite Reductase in Leaves of Barley Seedlings 1. Plant Physiol. 1989, 91, 1152–1156. [Google Scholar] [CrossRef]
- Stitt, M.; Müller, C.; Matt, P.; Gibon, Y.; Carillo, P.; Morcuende, R.; Scheible, W.; Krapp, A. Steps towards an Integrated View of Nitrogen Metabolism. J. Exp. Bot. 2002, 53, 959–970. [Google Scholar] [CrossRef]
- Solomonson, L.P.; Spehar, A.M. Model for the Regulation of Nitrate Assimilation. Nature 1977, 265, 373–375. [Google Scholar] [CrossRef]
- Yu, X.; Sukumaran, S.; Márton, L. Differential Expression of the Arabidopsis Nia1 andNia2 Genes1: Cytokinin-Induced Nitrate Reductase Activity Is Correlated With Increased Nia1 Transcription and mRNA Levels. Plant Physiol. 1998, 116, 1091–1096. [Google Scholar] [CrossRef]
- Kolbert, Z.; Bartha, B.; Erdei, L. Exogenous Auxin-Induced NO Synthesis Is Nitrate Reductase-Associated in Arabidopsis thaliana Root Primordia. J. Plant Physiol. 2008, 165, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.Q.; Crawford, N.M. Identification and Characterization of a Chlorate-Resistant Mutant of Arabidopsis thaliana with Mutations in Both Nitrate Reductase Structural Genes NIA1 and NIA2. Mol. Gen. Genet. MGG 1993, 239, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Jia, L.; Li, Y.; Smith, S.J.; Miller, A.J.; Shen, Q. Comparing Nitrate Storage and Remobilization in Two Rice Cultivars That Differ in Their Nitrogen Use Efficiency. J. Exp. Bot. 2007, 58, 1729–1740. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, Y.; Chen, G.; Zhang, A.; Yang, S.; Shang, L.; Wang, D.; Ruan, B.; Liu, C.; Jiang, H.; et al. The Indica Nitrate Reductase Gene OsNR2 Allele Enhances Rice Yield Potential and Nitrogen Use Efficiency. Nat. Commun. 2019, 10, 5207. [Google Scholar] [CrossRef] [PubMed]
- Costa-Broseta, Á.; Castillo, M.; León, J. Nitrite Reductase 1 Is a Target of Nitric Oxide-Mediated Post-Translational Modifications and Controls Nitrogen Flux and Growth in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 7270. [Google Scholar] [CrossRef]
- Yu, J.; Xuan, W.; Tian, Y.; Fan, L.; Sun, J.; Tang, W.; Chen, G.; Wang, B.; Liu, Y.; Wu, W.; et al. Enhanced OsNLP4-OsNiR Cascade Confers Nitrogen Use Efficiency by Promoting Tiller Number in Rice. Plant Biotechnol. J. 2021, 19, 167–176. [Google Scholar] [CrossRef]
- Cao, X.; Lu, X.; Xiong, J.; Li, J.; Xie, S. The Poncirus trifoliata (L.) Raf. NIN-Like Protein Transcription Factors Responses to Drought Stress and Bind the Nitrate-Responsive Cis-Element. Sci. Agric. Sin. 2018, 51, 3370–3378. [Google Scholar]
- Hirel, B.; Krapp, A. Nitrogen Utilization in Plants I Biological and Agronomic Importance. In Encyclopedia of Biochemistry; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Ghoshroy, S.; Binder, M.; Tartar, A.; Robertson, D.L. Molecular Evolution of Glutamine Synthetase II: Phylogenetic Evidence of a Non-Endosymbiotic Gene Transfer Event Early in Plant Evolution. BMC Evol. Biol. 2010, 10, 198. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, R.D.; Murray, A.J.S.; Lea, P.J. Inhibition of Photosynthesis in Barley with Decreased Levels of Chloroplastic Glutamine Synthetase Activity. J. Exp. Bot. 1987, 38, 1799–1809. [Google Scholar] [CrossRef]
- Wallsgrove, R.M.; Turner, J.C.; Hall, N.P.; Kendall, A.C.; Bright, S.W.J. Barley Mutants Lacking Chloroplast Glutamine Synthetase—Biochemical and Genetic Analysis. Plant Physiol. 1987, 83, 155–158. [Google Scholar] [CrossRef]
- Lal, S.K.; Mehta, S.; Raju, D.; Achary, V.M.M.; Venkatapuram, A.K.; Yadav, S.K.; Parmar, H.; Pandey, R.; Panditi, V.; Sheri, V.; et al. Concurrent Overexpression of Rice GS1;1 and GS2 Genes to Enhance the Nitrogen Use Efficiency (NUE) in Transgenic Rice. J. Plant Growth Regul. 2023, 42, 6699–6720. [Google Scholar] [CrossRef]
- Martin, A.; Lee, J.; Kichey, T.; Gerentes, D.; Zivy, M.; Tatout, C.; Dubois, F.; Balliau, T.; Valot, B.; Davanture, M.; et al. Two Cytosolic Glutamine Synthetase Isoforms of Maize Are Specifically Involved in the Control of Grain Production. Plant Cell 2006, 18, 3252–3274. [Google Scholar] [CrossRef]
- Li, Q.; Gao, Y.; Wang, K.; Feng, J.; Sun, S.; Lu, X.; Liu, Z.; Zhao, D.; Li, L.; Wang, D. Transcriptome Analysis of the Effects of Grafting Interstocks on Apple Rootstocks and Scions. Int. J. Mol. Sci. 2023, 24, 807. [Google Scholar] [CrossRef]
- van den Heuvel, R.H.H.; Ferrari, D.; Bossi, R.T.; Ravasio, S.; Curti, B.; Vanoni, M.A.; Florencio, F.J.; Mattevi, A. Structural Studies on the Synchronization of Catalytic Centers in Glutamate Synthase. J. Biol. Chem. 2002, 277, 24579–24583. [Google Scholar] [CrossRef][Green Version]
- Bowsher, C.G.; Lacey, A.E.; Hanke, G.T.; Clarkson, D.T.; Saker, L.R.; Stulen, I.; Emes, M.J. The Effect of Glc6P Uptake and Its Subsequent Oxidation within Pea Root Plastids on Nitrite Reduction and Glutamate Synthesis. J. Exp. Bot. 2007, 58, 1109–1118. [Google Scholar] [CrossRef][Green Version]
- Suzuki, A. Glutamate Synthase and Amino Acid Synthesis in Higher Plants. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2021; Volume 100, pp. 129–144. [Google Scholar][Green Version]
- Zeng, D.-D.; Qin, R.; Li, M.; Alamin, M.; Jin, X.-L.; Liu, Y.; Shi, C.-H. The Ferredoxin-Dependent Glutamate Synthase (OsFd-GOGAT) Participates in Leaf Senescence and the Nitrogen Remobilization in Rice. Mol. Genet. Genom. 2017, 292, 385–395. [Google Scholar] [CrossRef]
- Yamaya, T.; Obara, M.; Nakajima, H.; Sasaki, S.; Hayakawa, T.; Sato, T. Genetic Manipulation and Quantitative-trait Loci Mapping for Nitrogen Recycling in Rice. J. Exp. Bot. 2002, 53, 917–925. [Google Scholar] [CrossRef]
- Lyu, M.; Liu, J.; Xu, X.; Liu, C.; Qin, H.; Zhang, X.; Tian, G.; Jiang, H.; Jiang, Y.; Zhu, Z.; et al. Magnesium Alleviates Aluminum-Induced Growth Inhibition by Enhancing Antioxidant Enzyme Activity and Carbon–Nitrogen Metabolism in Apple Seedlings. Ecotoxicol. Environ. Saf. 2023, 249, 114421. [Google Scholar] [CrossRef]
- Su, H.; Li, L.; Ma, H.; Lyu, D.; Sun, J. Calcium Alleviates Temperature Stress by Regulating Nitrogen and Respiratory Metabolism in Malus baccata Roots. IJAB 2016, 18, 286–292. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; Van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R. High-Quality de Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Brunak, S.; von Heijne, G. Machine Learning Approaches for the Prediction of Signal Peptides and Other Protein Sorting Signals. Protein Eng. 1999, 12, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Hulo, N.; Sigrist, C.J.; Le Saux, V.; Langendijk-Genevaux, P.S.; Bordoli, L.; Gattiker, A.; De Castro, E.; Bucher, P.; Bairoch, A. Recent Improvements to the PROSITE Database. Nucleic Acids Res. 2004, 32, D134–D137. [Google Scholar] [CrossRef]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and Structure-Based Prediction of Eukaryotic Protein Phosphorylation Sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a Toolkit for Biologists Integrating Various Biological Data Handling Tools with a User-Friendly Interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, H.; Cui, T. Identification of a Strain Degrading Ammonia Nitrogen, Optimization of Ammonia Nitrogen Degradation Conditions, and Gene Expression of Key Degrading Enzyme Nitrite Reductase. Fermentation 2023, 9, 397. [Google Scholar] [CrossRef]
- Valderrama-Martín, J.M.; Ortigosa, F.; Ávila, C.; Cánovas, F.M.; Hirel, B.; Cantón, F.R.; Cañas, R.A. A Revised View on the Evolution of Glutamine Synthetase Isoenzymes in Plants. Plant J. 2022, 110, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Pageau, K.; Lelandais, M.; Grandjean, O.; Kronenberger, J.; Valadier, M.H.; Feraud, M.; Jouglet, T.; Suzuki, A. Glutamine Synthetase-Glutamate Synthase Pathway and Glutamate Dehydrogenase Play Distinct Roles in the Sink-Source Nitrogen Cycle in Tobacco. Plant Physiol. 2020, 140, 444–456. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).