Nitrogen Degradation Pathways in Actinomycetes: Key Components of Primary Metabolism Ensuring Survival in the Environment
Abstract
1. Introduction
1.1. Nitrogen Metabolism in Prokaryotes
1.2. Nitrogen Uptake and Nitrogen Assimilation in Actinobacteria
2. Molecular Mechanisms of Assimilation of Different Nitrogen Sources in Actinobacteria
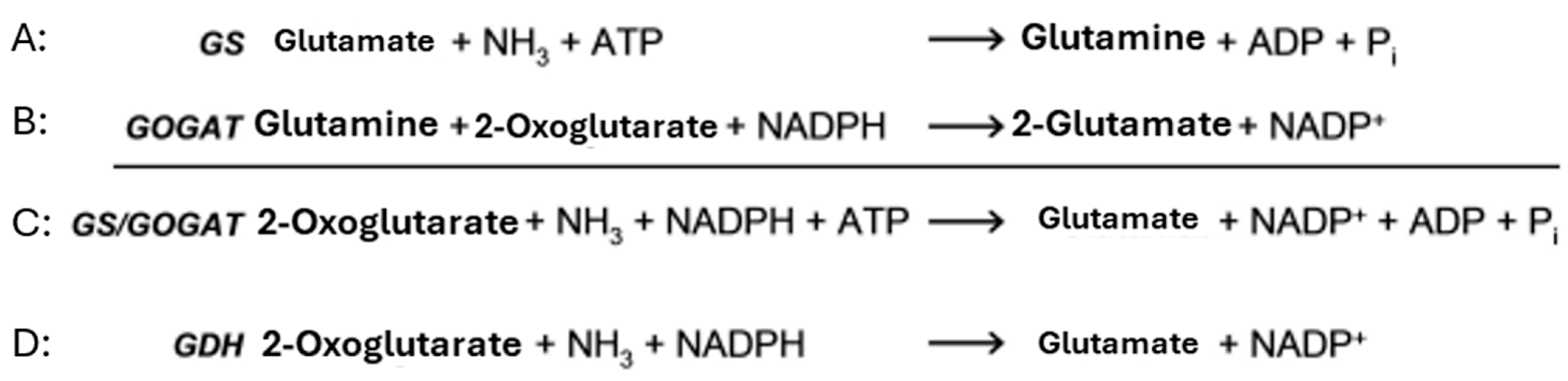
2.1. Nitrogen Assimilation in Actinomycetes: Ammonium Catabolism as Central Catabolic Route
2.2. Nitrogen Assimilation in Actinobacteria: Catabolism of Poor Nitrogen Sources
2.3. Assimilation of Amines in Actinomycetes
3. Nitrogen Assimilation in Actinobacteria: Transcriptional Regulation
3.1. Regulation of Central Pathways
3.2. Regulation of Catabolism of Amines in Actinomycetes
4. Summary and Discussion
5. Conclusions and Future Perspectives
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fuchs, G. (Ed.) Allgemeine Mikrobiologie, 11th ed.; Thieme: Stuttgart, Germany, 2022; ISBN 9783132434776. [Google Scholar]
- Aharonowitz, Y. Nitrogen metabolite regulation of antibiotic biosynthesis. Annu. Rev. Microbiol. 1980, 34, 209–233. [Google Scholar] [CrossRef]
- Reitzer, L.; Schneider, B.L. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 2001, 65, 422–444. [Google Scholar] [CrossRef]
- Ninfa, A.J.; Jiang, P.; Atkinson, M.R.; Peliska, J.A. Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr. Top. Cell. Regul. 2001, 36, 31–75. [Google Scholar]
- Hojati, Z.; Milne, C.; Harvey, B.; Gordon, L.; Borg, M.; Flett, F.; Wilkinson, B.; Sidebottom, P.J.; Rudd, B.A.; Hayes, M.A.; et al. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem. Biol. 2002, 9, 1175–1187. [Google Scholar] [CrossRef]
- Hopwood, D.A. The Leeuwenhoek Lecture, 1987: Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc. R. Soc. London Ser. B. Biol. Sci. 1988, 235, 121–138. [Google Scholar]
- Krysenko, S.; Wohlleben, W. Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity. Med. Sci. 2022, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Neidharth, F.C.; Reitzer, L.J. Regulation of Nitrogen Utilization Escherichia coli and Salmonella: Cellular and Molecular Biology; ASM Press: Washington, DC, USA, 1996; pp. 1344–1356. [Google Scholar]
- Jacoby, R.P.; Succurro, A.; Kopriva, S. Nitrogen Substrate Utilization in Three Rhizosphere Bacterial Strains Investigated Using Proteomics. Front. Microbiol. 2020, 11, 784. [Google Scholar] [CrossRef] [PubMed]
- Gouzy, A.; Poquet, Y.; Neyrolles, O. Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat. Rev. Microbiol. 2014, 12, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Rodríguez-García, A.; Liras, P. The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: Comparison in Streptomyces coelicolor and Streptomyces avermitilis. J. Antibiot. 2017, 70, 534–541. [Google Scholar] [CrossRef]
- Harper, C.; Hayward, D.; Wiid, I.; van Helden, P. Regulation of nitrogen metabolism in Mycobacterium tuberculosis: A comparison with mechanisms in Corynebacterium glutamicum and Streptomyces coelicolor. IUBMB Life 2008, 60, 643–650. [Google Scholar] [CrossRef]
- Martín, J.F.; Liras, P. The Balance Metabolism Safety Net: Integration of Stress Signals by Interacting Transcriptional Factors in Streptomyces and Related Actinobacteria. Front. Microbiol. 2020, 10, 3120. [Google Scholar] [CrossRef]
- Martín, J.F.; Sola-Landa, A.; Santos-Beneit, F.; Fernández-Martínez, L.T.; Prieto, C.; Rodríguez-García, A. Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb. Biotechnol. 2011, 4, 165–174. [Google Scholar] [CrossRef]
- Amon, J.; Titgemeyer, F.; Burkovski, A. Common patterns—Unique features: Nitrogen metabolism and regulation in Gram-positive bacteria. FEMS Microbiol. Rev. 2010, 34, 588–605. [Google Scholar] [CrossRef]
- Reuther, J.; Wohlleben, W. Nitrogen metabolism in Streptomyces coelicolor: Transcriptional and post-translational regulation. J. Mol. Microbiol. Biotechnol. 2007, 12, 139–146. [Google Scholar] [CrossRef]
- Rehm, N.; Burkovski, A. Engineering of nitrogen metabolism and its regulation in Corynebacterium glutamicum: Influence on amino acid pools and production. Appl. Microbiol. Biotechnol. 2011, 89, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Gtari, M.; Ghodhbane-Gtari, F.; Nouioui, I.; Beauchemin, N.; Tisa, L.S. Phylogenetic perspectives of nitrogen-fixing actinobacteria. Arch. Microbiol. 2012, 194, 3–11. [Google Scholar] [CrossRef]
- Zhang, M.; He, T.; Wu, P.; Wang, C.; Zheng, C. Recent advances in the nitrogen cycle involving actinomycetes: Current situation, prospect and challenge. Bioresour. Technol. 2025, 419, 132100. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Matthews, A.; Busche, T.; Bera, A.; Wohlleben, W. Poly- and Monoamine Metabolism in Streptomyces coelicolor: The New Role of Glutamine Synthetase-Like Enzymes in the Survival under Environmental Stress. Microb. Physiol. 2021, 31, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Romero-Rodríguez, A.; Maldonado-Carmona, N.; Ruiz-Villafán, B.; Koirala, N.; Rocha, D.; Sánchez, S. Interplay between carbon, nitrogen and phosphate utilization in the control of secondary metabolite production in Streptomyces. Antonie Van Leeuwenhoek 2018, 111, 761–781. [Google Scholar] [CrossRef]
- Makhoba, X.H.; Krysenko, S. Drug Target Validation in Polyamine Metabolism and Drug Discovery Advancements to Combat Tuberculosis. Future Pharmacol. 2025, 5, 32. [Google Scholar] [CrossRef]
- Magasanik, B. Genetic control of nitrogen assimilation in bacteria. Annu. Rev. Genet. 1982, 16, 135–168. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A. Forty years of genetics with Streptomyces: From in vivo through in vitro to in silico. Microbiology 1999, 145, 2183–2202. [Google Scholar] [CrossRef] [PubMed]
- Merrick, M.J.; Edwards, R.A. Nitrogen. control in bacteria. Microbiol. Rev. 1995, 59, 604–622. [Google Scholar] [CrossRef]
- Fischer, M.; Alderson, J.; van Keulen, G.; White, J.; Sawers, R.G. The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 2010, 156 Pt 10, 3166–3179. [Google Scholar] [CrossRef]
- Voelker, F.; Altaba, S. Nitrogen source governs the patterns of growth and pristinamycin production in ‘Streptomyces pristinaespiralis’. Microbiology 2001, 147 Pt 9, 2447–2459. [Google Scholar] [CrossRef]
- Wray, L.V., Jr.; Fisher, S.H. Cloning and nucleotide sequence of the Streptomyces coelicolor gene encoding glutamine synthetase. Gene 1988, 71, 247–256. [Google Scholar] [CrossRef]
- Wray, L.V., Jr.; Fisher, S.H. The Streptomyces coelicolor glnR gene encodes a protein similar to other bacterial response regulators. Gene 1993, 130, 145–150. [Google Scholar] [CrossRef]
- Wray, L.V.; Atkinson, M.R.; Fisher, S.H. Identification and cloning of the glnR locus, which is required for transcription of the glnA gene in Streptomyces coelicolor A3(2). J. Bacteriol. 1991, 173, 7351–7360. [Google Scholar] [CrossRef]
- Fisher, S.H. Glutamate synthesis in Streptomyces coelicolor. J. Bacteriol. 1989, 171, 2372–2377. [Google Scholar] [CrossRef]
- Streicher, S.L.; Tyler, B. Regulation of glutamine synthetase activity by adenylylation in the Gram-positive bacterium Streptomyces cattleya. Proc. Natl. Acad. Sci. USA 1981, 78, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Fink, D.; Falke, D.; Wohlleben, W.; Engels, A. Nitrogen metabolism in Streptomyces coelicolor A3(2): Modification of glutamine synthetase I by an adenylyltransferase. Microbiology 1999, 145 Pt 9, 2313–2322. [Google Scholar] [CrossRef]
- Nolden, L.; Farwick, M.; Kramer RBurkovski, A. Glutamine synthetases of Corynebacterium glutamicum: Transcriptional control and regulation of activity. FEMS Microbiol. Lett. 2001, 201, 91–98. [Google Scholar] [CrossRef]
- Williams, K.J.; Bennett, M.H.; Barton, G.R.; Jenkins, V.A.; Robertson, B.D. Adenylylation of mycobacterial Glnk (PII) protein is induced by nitrogen limitation. Tuberculosis 2013, 93, 198–206. [Google Scholar] [CrossRef][Green Version]
- Rehm, N.; Buchinger, S.; Strosser, J.; Dotzauer, A.; Walter, B.; Hans, S.; Bathe, B.; Schomburg, D.; Kramer RBurkovski, A. Impact of adenylyltransferase GlnE on nitrogen starvation response in Corynebacterium glutamicum. J. Biotechnol. 2010, 145, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Darrow, R.A.; Knotts, R.R. Two forms of glutamine synthetase in free-living root-nodule bacteria. Biochem. Biophys. Res. Commun. 1977, 78, 554–559. [Google Scholar] [CrossRef]
- Ghoshroy, S.; Binder, M.; Tartar, A.; Robertson, D.L. Molecular evolution of glutamine synthetase II: Phylogenetic evidence of a non-endosymbiotic gene transfer event early in plant evolution. BMC Evol. Biol. 2010, 10, 198. [Google Scholar] [CrossRef] [PubMed]
- Edmands, J.; Noridge, N.A.; Benson, D.R. The actinorhizal root-nodule symbiont Frankia sp. strain CpI1 has two glutamine synthetases. Proc. Natl. Acad. Sci. USA 1987, 84, 6126–6130. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, I.; Hillemann, D.; Pühler, A.; Strauch, E.; Wohlleben, W. Overexpression of a Streptomyces viridochromogenes gene (glnII) encoding a glutamine synthetase similar to those of eucaryotes confers resistance against the antibiotic phosphinothricyl-alanyl-alanine. J. Bacteriol. 1990, 172, 5326–5334. [Google Scholar] [CrossRef]
- Kumada, Y.; Benson, D.R.; Hillemann, D.; Hosted, T.J.; Rochefort, D.A.; Thompson, C.J.; Wohlleben, W.; Tateno, Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc. Natl. Acad. Sci. USA 1993, 90, 3009–3013. [Google Scholar] [CrossRef]
- Hillemann, D.; Dammann, T.; Hillemann, A.; Wohlleben, W. Genetic and biochemical characterization of the two glutamine synthetases GSI and GSII of the phosphinothricyl-alanyl-alanine producer, Streptomyces viridochromogenes Tü494. J. Gen. Microbiol. 1993, 139, 1773–1783. [Google Scholar] [CrossRef][Green Version]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete Genome Sequence of the Model Actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Harth, G.; Masleša-Galić, S.; Tullius, M.V.; Horwitz, M.A.A. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 2005, 58, 1157–1172. [Google Scholar] [CrossRef]
- Fink, D.; Weißschuh, N.; Reuther, J.; Wohlleben, W.; Engels, A. Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 2002, 46, 331–347. [Google Scholar] [CrossRef]
- Rexer, H.U.; Schäberle, T.; Wohlleben, W.; Engels, A. Investigation of the functional properties and regulation of three glutamine synthetase-like genes in Streptomyces coelicolor A3(2). Arch. Microbiol. 2006, 186, 447–458. [Google Scholar] [CrossRef]
- Krysenko, S.; Okoniewski, N.; Kulik, A.; Matthews, A.; Grimpo, J.; Wohlleben, W.; Bera, A. Gamma-Glutamylpolyamine Synthetase GlnA3 is involved in the first step of polyamine degradation pathway in Streptomyces coelicolor M145. Front. Microbiol. 2017, 8, 726. [Google Scholar] [CrossRef] [PubMed]
- Takeo, M.; Ohara, A.; Sakae, S.; Okamoto, Y.; Kitamura, C.; Kato, D.; Negoro, S. Function of a glutamine synthetase-like protein in bacterial aniline oxidation via γ-glutamylanilide. J. Bacteriol. 2013, 195, 4406–4414. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, V.A.; Barton, G.R.; Robertson, B.D.; Williams, K.J. Genome wide analysis of the complete GlnR nitrogen-response regulon in Mycobacterium smegmatis. BMC Genom. 2013, 14, 301. [Google Scholar] [CrossRef] [PubMed]
- Jeßberger, N.; Lu, Y.; Amon, J.; Titgemeyer, F.; Sonnewald, S.; Reid, S.; Burkovski, A. Nitrogen starvation-induced transcriptome alterations and influence of transcription regulator mutants in Mycobacterium smegmatis. BMC Res. Notes 2013, 6, 482. [Google Scholar] [CrossRef]
- Deshpande, K.L.; Katze, J.R.; Kane, J.F. Effect of glutamine on enzymes of nitrogen metabolism in Bacillus subtilis. J. Bacteriol. 1981, 145, 768–774. [Google Scholar] [CrossRef]
- Chipeva, V.; Dumanova, E.; Todorov, T.; Ivanova, I. Impact of nitrogen assimilation on regulation of antibiotic production in Streptomyces hygroscopicus 155. Antibiot. Khimioterapiia Antibiot. Chemoterapy 1991, 36, 5–8. [Google Scholar]
- Bascaran, V.; Hardisson, C.; Brana, A.F. Regulation of nitrogen catabolic enzymes in Streptomyces clavuligerus. J. Gen. Microbiol. 1989, 135, 2465–2474. [Google Scholar]
- Allaway, D.; Lodwig, E.; Crompton, L.A.; Wood, M.; Parsons, R.; Wheeler, T.R.; Poole, P.S. Identification of alaninedehydrogenase and its role in mixed secretion of ammonium andalanine by pea bacteroids. Mol. Microbiol. 2000, 36, 508–515. [Google Scholar] [CrossRef]
- Shapiro, S.; Vining, L.C. Nitrogen. metabolism and chloramphenicol production in Streptomyces venezuelae. Can. J. Microbiol. 1983, 29, 1706–1714. [Google Scholar] [CrossRef]
- Hudson, R.C.; Daniel, R.M. L-glutamate dehydrogenases: Distribution, properties and mechanism. Comp. Biochem. Physiol. Part B Comp. Biochem. 1993, 106, 767–792. [Google Scholar] [CrossRef] [PubMed]
- Tyler, B. Regulation of the assimilation of nitrogen compounds. Annu. Rev. Biochem. 1978, 47, 1127–1162. [Google Scholar] [CrossRef]
- Kawakami, R.; Sakuraba, H.; Ohshima, T. Gene cloning and characterization of the very large NAD-dependent l-glutamate dehydrogenase from the psychrophile Janthinobacterium lividum, isolated from cold soil. J. Bacteriol. 2007, 189, 5626–5633. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, B.G. NAD(+)-specific glutamate dehydrogenase (EC.1.4.1.2) in Streptomyces coelicolor; in vivo characterization and the implication for nutrient-dependent secondary metabolism. Appl. Microbiol. Biotechnol. 2016, 100, 5527–5536. [Google Scholar] [CrossRef]
- Engel, P.C. Glutamate dehydrogenases: The why and how of coenzyme specificity. Neurochem. Res. 2014, 39, 426–432. [Google Scholar] [CrossRef]
- Plaitakis, A.; Kalef-Ezra, E.; Kotzamani, D.; Zaganas, I.; Spanaki, C. The Glutamate Dehydrogenase Pathway and Its Roles in Cell and Tissue Biology in Health and Disease. Biology 2017, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A.; Chater, K.F.; Bibb, M.J. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology 1995, 28, 65–102. [Google Scholar]
- Goddard, A.D.; Moir, J.W.B.; Richardson, D.J.; Ferguson, S.J. Interdependence of two NarK domains in a fused nitrate/nitrite transporter. Mol. Microbiol. 2008, 70, 667–681. [Google Scholar] [CrossRef]
- Goddard, A.D.; Bali, S.; Mavridou, D.A.; Luque-Almagro, V.M.; Gates, A.J.; Roldán, M.D.; Newstead, S.; Richardson, D.J.; Ferguson, S.J. The Paracoccus denitrificans NarK-like nitrate and nitrite transporters-probing nitrate uptake and nitrate/nitrite exchange mechanisms. Mol. Microbiol. 2017, 103, 117–133. [Google Scholar] [CrossRef]
- Stolz, J.F.; Basu, P. Evolution of nitrate reductase: Molecular and structural variations on a common function. Chembiochem 2002, 3, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Sparacino-Watkins, C.; Stolz, J.F.; Basu, P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014, 43, 676–706. [Google Scholar] [CrossRef]
- Hsiao, N.H.; Kirby, R. Comparative genomics of Streptomyces avermitilis, Streptomyces cattleya, Streptomyces maritimus and Kitasatospora aureofaciens using a Streptomyces coelicolor microarray system. Antonie Van Leeuwenhoek 2008, 93, 1–25. [Google Scholar] [CrossRef][Green Version]
- Fischer, M.; Falke, D.; Sawers, R.G. A respiratory nitrate reductase active exclusively in resting spores of the obligate aerobe Streptomyces coelicolor A3(2). Mol. Microbiol. 2013, 89, 1259–1273. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Falke, D.; Pawlik, T.; Sawers, R.G. Oxygen-dependent control of respiratory nitrate reduction in mycelium of Streptomyces coelicolor A3(2). J. Bacteriol. 2014, 196, 4152–4162. [Google Scholar] [CrossRef]
- Richardson, D.J.; Berks, B.C.; Russell, D.A.; Spiro, S.; Taylor, C.J. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. CMLS 2001, 58, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. MMBR 1997, 61, 533–616. [Google Scholar] [CrossRef] [PubMed]
- Higgins, S.A.; Welsh, A.; Orellana, L.H.; Konstantinidis, K.T.; Chee-Sanford, J.C.; Sanford, R.A.; Schadt, C.W.; Löffler, F.E. Detection and Diversity of Fungal Nitric Oxide Reductase Genes. (p450nor) in Agricultural Soils. Appl. Environ. Microbiol. 2016, 82, 2919–2928. [Google Scholar] [CrossRef]
- Richter, C.D.; Allen, J.W.; Higham, C.W.; Koppenhofer, A.; Zajicek, R.S.; Watmough, N.J.; Ferguson, S.J. Cytochrome cd1, reductive activation and kinetic analysis of a multifunctional respiratory enzyme. J. Biol. Chem. 2002, 277, 3093–3100. [Google Scholar] [CrossRef]
- Wang, H.; Gunsalus, R.P. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J. Bacteriol. 2000, 182, 5813–5822. [Google Scholar] [CrossRef]
- Besson, S.; Almeida, M.G.; Silveira, C.M. Nitrite reduction in bacteria: A comprehensive view of nitrite reductases. Coord. Chem. Rev. 2022, 464, 214560. [Google Scholar] [CrossRef]
- Yukioka, Y.; Tanahashi, T.; Shida, K.; Oguchi, H.; Ogawa, S.; Saito, C.; Yajima, S.; Ito, S.; Ohsawa, K.; Shoun, H.; et al. A role of nitrite reductase (NirBD) for NO homeostatic regulation in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2017, 364, fnw241. [Google Scholar] [CrossRef]
- Tiffert, Y.; Supra, P.; Wurm, R.; Wohlleben, W.; Wagner, R.; Reuther, J. The Streptomyces coelicolor GlnR regulon: Identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 2008, 67, 861–880. [Google Scholar] [CrossRef]
- Tiffert, Y.; Franz-Wachtel, M.; Fladerer, C.; Nordheim, A.; Reuther, J.; Wohlleben, W.; Mast, Y. Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl. Microbiol. Biotechnol. 2011, 89, 1149–1159. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Reuther, J.; Bera, A.; Wohlleben, W.; Mast, Y. A novel GlnR target gene, nnaR, is involved in nitrate/nitrite assimilation in Streptomyces coelicolor. Microbiology 2012, 158, 1172–1182. [Google Scholar] [CrossRef]
- Jakoby, M.; Nolden, L.; Meier-Wagner, J.; Krämer, R.; Burkovski, A. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol. Microbiol. 2000, 37, 964–977. [Google Scholar] [CrossRef] [PubMed]
- van Heeswijk, W.C.; Hoving, S.; Molenaar, D.; Stegeman, B.; Kahn, D.; Westerhoff, H.V.A. alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol. Microbiol. 1996, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, A.; Fink, D.; Gust, B.; Rexer, H.; Scheel, B.; Chater, K.; Wohlleben, W.; Engels, A. The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 2002, 46, 319–330. [Google Scholar] [CrossRef]
- Ensinck, D.; Gerhardt, E.C.M.; Rollan, L.; Huergo, L.F.; Gramajo, H.; Diacovich, L. The PII protein interacts with the Amt ammonium transport and modulates nitrate/nitrite assimilation in mycobacteria. Front. Microbiol. 2024, 15, 1366111. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, M.R.; Ninfa, A.J. Mutational analysis of the bacterial signal-transducing protein kinase/phosphatase nitrogen regulator II (NRII or NtrB). J. Bacteriol. 1993, 175, 7016–7023. [Google Scholar] [CrossRef][Green Version]
- Atkinson, M.R.; Ninfa, A.J. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 1998, 29, 431–447. [Google Scholar] [CrossRef]
- Atkinson, M.R.; Ninfa, A.J. Characterization of the GlnK protein of Escherichia coli. Mol. Microbiol. 1999, 32, 301–313. [Google Scholar] [CrossRef]
- Radchenko, M.V.; Thornton, J.; Merrick, M. Control of AmtB-GlnK complex formation by intracellular levels of ATP, ADP, and 2-oxoglutarate. J. Biol. Chem. 2010, 285, 31037–31045. [Google Scholar] [CrossRef] [PubMed]
- Radchenko, M.V.; Thornton, J.; Merrick, M. P(II) signal transduction proteins are ATPases whose activity is regulated by 2-oxoglutarate. Proc. Natl. Acad. Sci. USA 2013, 110, 12948–12953. [Google Scholar] [CrossRef]
- Forchhammer, K. Glutamine signalling in bacteria. Front Biosci. 2007, 12, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.H.; Sonenshein, A.L. Control of carbon and nitrogen metabolism in Bacillus subtilis. Annu. Rev. Microbiol. 1991, 45, 107–135. [Google Scholar] [CrossRef]
- Strösser, J.; Ludke, A.; Schaffer, S.; Kramer, R.; Burkovski, A. Regulation of GlnK activity: Modification, membrane sequestration and proteolysis as regulatory principles in the network of nitrogen control in Corynebacterium glutamicum. Mol. Microbiol. 2004, 54, 132–147. [Google Scholar] [CrossRef]
- Beckers, G.; Strösser, J.; Hildebrandt, U.; Kalinowski, J.; Farwick, M.; Krämer, R.; Burkovski, A. Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: Mechanism and characterization of the AmtR regulon. Mol. Microbiol. 2005, 58, 580–595. [Google Scholar] [CrossRef]
- Heinrich, A.; Woyda, K.; Brauburger, K.; Meiss, G.; Detsch, C.; Stulke, J.; Forchhammer, K. Interaction of the membrane-bound GlnK-AmtB complex with the master regulator of nitrogen metabolism TnrA in Bacillus subtilis. J. Biol. Chem. 2006, 281, 34909–34917. [Google Scholar] [CrossRef]
- Michael, A.J. Polyamine function in archaea and bacteria. J. Biol. Chem. 2018, 293, 18693–18701. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.S. A Guide to Polyamines; Oxford University Press: Oxford, UK, 1998. [Google Scholar]
- Igarashi, K.; Kashiwagi, K. Polyamines: Mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 2000, 271, 559–564. [Google Scholar] [CrossRef]
- Kusano, T.; Suzuki, H. (Eds.) Polyamines: A Universal Molecular Nexus for Growth, Survival, and Specialized Metabolism; Springer: Tokyo, Japan, 2015. [Google Scholar]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef]
- Knaak, J.B.; Leung, H.-W.; Stott, W.T.; Busch, J.; Bilsky, J. Toxicology of mono-, di-, and triethanolamine. Rev. Environ. Contam. Toxicol. 1997, 149, 1–86. [Google Scholar] [CrossRef]
- Burrell, M.; Hanfrey, C.C.; Kinch, L.N.; Elliot, K.A.; Michael, J.A. Evolution of a novel lysine decarboxylase in siderophore biosynthesis. Mol. Microbiol. 2012, 86, 485–499. [Google Scholar] [CrossRef]
- Kurihara, S.; Oda, S.; Tsuboi, Y.; Kim, H.G.; Oshida, M.; Kumagai, H.; Suzuki, H. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J. Biol. Chem. 2008, 283, 19981–19990. [Google Scholar] [CrossRef]
- Kurihara, S.; Sakai, Y.; Suzuki, H.; Muth, A.; Ot, P.; Rather, P.N. Putrescine importer PlaP contributes to swarming motility and urothelial cell invasion in Proteus mirabilis. J. Biol. Chem. 2013, 288, 15668–15676. [Google Scholar] [CrossRef]
- Kurihara, S.; Oda, S.; Kato, K.; Kim, H.G.; Koyanagi, T.; Kumagai, H.; Suzuki, H. A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J. Biol. Chem. 2005, 280, 4602–4608. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Lu, C.D. Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2006, 50, 1615–1622. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Wohlleben, W. Role of Carbon, Nitrogen, Phosphate and Sulfur Metabolism in Secondary Metabolism Precursor Supply in Streptomyces spp. Microorganisms 2024, 12, 1571. [Google Scholar] [CrossRef] [PubMed]
- Perez-Redondo, R.; Rodriguez-Garcia, A.; Botas, A.; Santamarta, I.; Martin, J.F.; Liras, P. ArgR of Streptomyces coelicolor is a versatile regulator. PLoS ONE 2012, 7, e32697. [Google Scholar] [CrossRef]
- Krysenko, S.; Matthews, A.; Okoniewski, N.; Kulik, A.; Girbas, M.G.; Tsypik, O.; Meyners, C.S.; Hausch, F.; Wohlleben, W.; Bera, A. Initial metabolic step of a novel ethanolamine utilization pathway and its regulation in Streptomyces coelicolor M145. mBio 2019, 10, e00326-19. [Google Scholar] [CrossRef]
- San Francisco, B.; Zhang, X.; Whalen, K.; Gerlt, K. A novel pathway for bacterial ethanolamine metabolism. FASEB J. 2015, 29, 573-45. [Google Scholar] [CrossRef]
- Gerlt, J.A. Tools and strategies for discovering novel enzymes and metabolic pathways. Perspect. Sci. 2016, 9, 24–32. [Google Scholar] [CrossRef]
- Amon, J.; Bräu, T.; Grimrath, A.; Hänßler, E.; Hasselt, K.; Höller, M.; Jeßberger, N.; Ott, L.; Szököl, J.; Titgemeyer, F.; et al. Nitrogen control in Mycobacterium smegmatis: Nitrogen-dependent expression of ammonium transport and assimilation proteins depends on OmpR-type regulator GlnR. J. Bacteriol. 2008, 190, 7108–7116. [Google Scholar] [CrossRef]
- Lin, W.; Wang, Y.; Han, X.; Zhang, Z.; Wang, C.; Wang, J.; Yang, H.; Lu, Y.; Jiang, W.; Zhao, G.-P.; et al. Atypical OmpR/PhoB subfamily response regulator GlnR of actinomycetes functions as a homodimer, stabilized by the unphosphorylated conserved Asp-focused charge interactions. J. Biol. Chem. 2014, 289, 15413–15425. [Google Scholar] [CrossRef]
- Muhl, D.; Jessberger, N.; Hasselt, K.; Jardin, C.; Sticht, H.; Burkovski, A. DNA binding by Corynebacterium glutamicum TetR-type transcription regulator AmtR. BMC Mol. Biol. 2009, 10, 73. [Google Scholar] [CrossRef]
- Amin, R.; Franz-Wachtel, M.; Tiffert, Y.; Heberer, M.; Meky, M.; Ahmed, Y.; Matthews, A.; Krysenko, S.; Jakobi, M.; Hinder, M.; et al. Post-translational Serine/Threonine Phosphorylation and Lysine Acetylation: A Novel Regulatory Aspect of the Global Nitrogen Response Regulator GlnR in S. coelicolor M145. Front. Mol. Biosci. 2016, 3, 38. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Okoniewski, N.; Nentwich, M.; Matthews, A.; Bäuerle, M.; Zinser, A.; Busche, T.; Kulik, A.; Gursch, S.; Kemeny, A.; et al. A Second Gamma-Glutamylpolyamine Synthetase, GlnA2, Is Involved in Polyamine Catabolism in Streptomyces coelicolor. Int. J. Mol. Sci. 2022, 23, 3752. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.L.; Liao, C.H.; Huang, G.; Zhou, Y.; Rigali, S.; Zhang, B.; Ye, B.C. GlnR-mediated regulation of nitrogen metabolism in the actinomycete Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 2014, 98, 7935–7948. [Google Scholar] [CrossRef]
- Atkinson, M.R.; Blauwkamp, T.A.; Bondarenko, V.; Studitsky, V.; Ninfa, A.J. Activation of the glnA, glnK, and nac promoters as Escherichia coli undergoes the transition from nitrogen excess growth to nitrogen starvation. J. Bacteriol. 2002, 184, 5358–5363. [Google Scholar] [CrossRef]
- Atkinson, M.R.; Kamberov, E.S.; Weiss, R.L.; Ninfa, A.J. Reversible uridylylation of the Escherichia coli PII signal transduction protein regulates its ability to stimulate the dephosphorylation of the transcription factor nitrogen regulator I (NRI or NtrC). J. Biol. Chem. 1994, 269, 28288–28293. [Google Scholar] [CrossRef]
- Reitzer, L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 2003, 57, 155–176. [Google Scholar] [CrossRef]
- Schulz, S.; Sletta, H.; Fløgstad Degnes, K.; Krysenko, S.; Williams, A.; Olsen, S.M.; Vernstad, K.; Mitulski, A.; Wohlleben, W. Optimization of FK-506 production in Streptomyces tsukubaensis by modulation of Crp-mediated regulation. Appl. Microbiol. Biotechnol. 2023, 107, 2871–2886. [Google Scholar] [CrossRef]
- Stewart, V. Dual interacting two-component regulatory systems mediate nitrate- and nitrite-regulated gene expression in Escherichia coli. Res. Microbiol. 1994, 145, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Zalkin, H.; Smith, J.L. Enzymes utilizing glutamine as an amide donor. Adv. Enzymol. Relat. Areas Mol. Biol. 1998, 72, 87–144. [Google Scholar]
- Wulandari, D.A.; Hartati, Y.W.; Ibrahim, A.U.; Pitaloka, D.A.E. Irkham Multidrug-resistant tuberculosis. Clin. Chim. Acta Int. J. Clin. Chem. 2024, 559, 119701. [Google Scholar] [CrossRef]
- Yan, L.; Niu, X.; Liang, K.; Guan, F.; Yu, X.; Ye, Z.; Huang, M.; Liang, H.; Zhong, X.; Zeng, J. Assessing the Role of Polyamine Metabolites in Blood and the DNA Methylation of Mycobacterium Tuberculosis in Patients with Multidrug-Resistant Tuberculosis. Int. J. Med. Sci. 2025, 22, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Baral, B.; Mozafari, M.R. Strategic Moves of “Superbugs” Against Available Chemical Scaffolds: Signaling, Regulation, and Challenges. ACS Pharmacol. Transl. Sci. 2020, 3, 373–400. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Lopez, M.; Meyners, C.; Purder, P.L.; Zinser, A.; Hausch, F.; Wohlleben, W. A novel synthetic inhibitor of polyamine utilization in Streptomyces coelicolor. FEMS Microbiol. Lett. 2023, 370, fnad096. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Emani, C.S.; Bäuerle, M.; Oswald, M.; Kulik, A.; Meyners, C.; Hillemann, D.; Merker, M.; Prosser, G.; Wohlers, I.; et al. GlnA3Mt is able to glutamylate spermine but it is not essential for the detoxification of spermine in Mycobacterium tuberculosis. J. Bacteriol. 2025, 207, e0043924. [Google Scholar] [CrossRef] [PubMed]



| Annotated Function | Homologue in S. coelicolor | Homologue in M. tuberculosis |
|---|---|---|
| glnA4 gamma–glutamylethanolamide synthetase | SCO1613 | Rv2860c |
| 2,4-Diaminobutyric acid Acetyltransferase | SCO1864 | Rv3225c |
| glnA (GSI) glutamine synthetase I | SCO2198 | Rv2220 |
| glnII (GSII) glutamine synthetase II | SCO2210 | - |
| glnRII Response Regulator | SCO2213 | Rv2884 |
| glnE adenylyltransferase | SCO2234 | Rv2221c |
| glnA2 gamma–glutamylpolyamine synthetase | SCO2241 | Rv2222c |
| glnR Response Regulator | SCO4159 | Rv0818 |
| gdhA Glutamate dehydrogenase | SCO4683 | Rv3726 |
| ureA, B, C α-, β-, γ-subunits of urease | SCO5525-6 | Rv1848-50 |
| amtB ammonium transporter | SCO5583 | Rv2920c |
| gluB glutamate transporter | SCO5776 | Rv2919c |
| gltB glutamate synthase | SCO1977 | Rv3859c |
| nirB2 nitrate reductase | SCO2486 | - |
| nirB nitrate reductase | SCO2487 | Rv0252 |
| nirD nitrate reductase | SCO2488 | Rv0253 |
| nnaR Transcriptional regulator of nitrate/nitrite assimilation | SCO2958 | Rv0260c |
| narK Nitrate Nitrite transporter | SCO2959 | Rv2329c |
| narGHIJ respiratory nitrate reductase | SCO6532-5 | Rv1161-4 |
| glnA3 gamma–glutamylpolyamine synthetase | SCO6962 | Rv1878 |
| narB nitrate reductase | SCO7374 | - |
| nasA nitrate reductase | SCO2473 | - |
| narK2 Nitrate Nitrite transporter | SCO0213 | Rv1737c |
| Annotated Function | Homologue in E. coli | Homologue in P. aeruginosa | Homologue in S. coelicolor | Homologue in M. tuberculosis |
|---|---|---|---|---|
| Polyamine ABC transporter ATP-binding protein PotA-like | PotA (b1126)/YdcT (b1441) | PAO603/PAO326 | SCO3453 | Rv2397c |
| Polyamine ABC transporter ATP-binding protein PotC-like | PotC (b1124)/YdcV (b1443) | PAO324/PotC (PA3609) | SCO3454 | Rv1237 |
| Polyamine ABC transporter protein | PotB (b1125)/YdcU (b1442) | PotB (PA0205)/PA3252 | SCO3455 | Rv2040c |
| Polyamine ABC transporter protein—substrate binding protein | YnjB (b1754) | PA0203 | SCO3456 | Rv3869 |
| Amino acid/polyamine permease | PuuP (b1296)/PlaP (b2014) | PA5510 | SCO5057 | Rv3253c |
| Lysine/ornithine decarboxylase-like enzyme | - | - | SCO5651 | Rv1205 |
| Pyruvate-polyamine aminotransferase | PatA (b3073) | SpuC (PA0299) | SCO5655 | Rv3329 |
| Lrp/AsnC family transcriptional regulator | - | - | SCO5656 | Rv2779c |
| γ-aminobutyraldehyde or γ-glutamyl-γ-amino-butyraldehyde dehydrogenase | PatD (b1444)/PuuC (b1300) | BetB (PA5373)/PAO219 | SCO5657 | Rv0458 |
| Polyamine-binding lipoprotein | PotF (b0854) | SpuD (PA0300) | SCO5658 | Rv3484 |
| γ-aminobutyraldehyde dehydrogenase or 4-guanidino-butyraldehyde dehydrogenase | PatD (b1444) PuuC (b1300) | PauC/KauB (PA5312) | SCO5666 | Rv2858c |
| Polyamine ABC transporter substrate-binding protein | PotF (b0854) | SpuE (PA0301) | SCO5667 | - |
| Polyamine ABC transporter substrate-binding protein | PotG (b0855) | SpuF (PA0302) | SCO5668 | Rv2397c |
| Polyamine ABC-transporter integral membrane protein | PotH (b0856) | SpuG (PA0303) | SCO5669 | Rv2399c |
| Polyamine ABC-transporter integral membrane protein | PotI (b0857) | SpuH (PA0304) | SCO5670 | Rv2398c |
| γ-glutamyl-polyamine oxidoreductase | PuuB (b1301) | PauB3 (PA2776) | SCO5671 | Rv3742c |
| γ-aminobutyrate aminotransferase gabT-like or puuE-like | GabT (b2662)/PuuE (b1302) | GabT (PA266) | SCO5676 | Rv2589 |
| Succinate-semialdehyde dehydrogenase gabD-like | GabD (b2661) | GabD (PA0265) | SCO5679 | Rv0223c |
| Amino acids/polyamine permease | PuuP (b1296) | PA5510/PAO322 | SCO5977 | Rv2320c |
| Hydrolase | - | - | SCO6960 | Rv0193c |
| Amidohydrolase | - | - | SCO6961 | Rv1879 |
| γ-glutamyl-polyamine synthetase | PuuA (b1297) | PauA7 (PA5508)/SpuI (PA0296) | SCO6962 | Rv1878 |
| γ-glutamyl ethanolamine synthetase/ethanolamine γ-glutamylase | - | - | SCO1613 | Rv2860c |
| γ-glutamyl ethanolamine dehydrogenase/iron-dependent dehydrogenase | - | - | SCO1611 | Rv1941 |
| γ-glutamyl aldehyde dehydrogenase | - | - | SCO1612 | Rv2858c |
| γ-glutamyl glycine amidohydrolase /formylglutamate amidohydrolase | - | - | SCO1615 | Rv2859c |
| Regulator Name | Function | Reference |
|---|---|---|
| GlnR | Central regulator of nitrogen metabolism regulating glnA, glnII, gdhA, nirB, ureA, and amtB-glnK-glnD | [78,116,117,118] |
| GlnRII | A GlnR homologue that recognizes glnA, amtB, and glnII | [45] |
| Crp | Regulates the interplay of primary and secondary metabolism, activating glnA, glnII, and amtB-glnK-glnD | [119] |
| ArgR | Controls the expression of glnR in response to nutrient stress stimuli | [7,105] |
| PhoP | Represses the amtB-glnK-glnD operon and glnA, glnII, and glnR under conditions of phosphate limitation | [7,105] |
| AfsR | Controls expression of glnR in response to unknown nutrient stress stimulus | [7,105] |
| AmtR | Regulates key genes involved in N metabolism, such as glnA, gltB, and amtB | [50] |
| AfsQ1 | Required for regulation of carbon, nitrogen, and phosphate metabolism in the presence of glutamate | [7,105] |
| EpuRI | Regulates genes for ethanolamine utilization glnA4, sco1612, sco1611, and sco1610 | [107] |
| EpuRII | Regulates genes for polyamine utilization glnA3, sco5676, sco5977, and sco6960 | [114] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krysenko, S. Nitrogen Degradation Pathways in Actinomycetes: Key Components of Primary Metabolism Ensuring Survival in the Environment. Nitrogen 2025, 6, 107. https://doi.org/10.3390/nitrogen6040107
Krysenko S. Nitrogen Degradation Pathways in Actinomycetes: Key Components of Primary Metabolism Ensuring Survival in the Environment. Nitrogen. 2025; 6(4):107. https://doi.org/10.3390/nitrogen6040107
Chicago/Turabian StyleKrysenko, Sergii. 2025. "Nitrogen Degradation Pathways in Actinomycetes: Key Components of Primary Metabolism Ensuring Survival in the Environment" Nitrogen 6, no. 4: 107. https://doi.org/10.3390/nitrogen6040107
APA StyleKrysenko, S. (2025). Nitrogen Degradation Pathways in Actinomycetes: Key Components of Primary Metabolism Ensuring Survival in the Environment. Nitrogen, 6(4), 107. https://doi.org/10.3390/nitrogen6040107






