Abstract
Bark beetle-associated bacteria from the sub-boreal and boreal forests of northern Canada represent a largely unexplored source of bioactive natural products. This study aims to investigate the chemical potential of bacteria isolated from Dendroctonus ponderosae, Dendroctonus rufipennis, Dendroctonus pseudotsugae, and Ips perturbatus by focusing on nitrogen-containing secondary metabolites. Genomic analyses of the bacterial isolates identified diverse biosynthetic gene clusters (BGCs), including nonribosomal peptides (NRPs), NRPS-PKS hybrids, and ribosomally synthesized and post-translationally modified peptides (RiPPs), many of which exhibit low sequence homology, suggesting potential for novel bioactive compounds. Nitrogen-15 NMR spectroscopy was employed to detect nitrogen-containing functional groups in crude extracts, revealing distinct signals for amides, amines, and nitrogen heterocycles. The combination of BGC predictions and NMR data highlighted the genetic and chemical diversity of these bacteria and underscored the potential for discovering novel nitrogen-rich metabolites. These findings provide a foundation for further exploration of bioactive natural products with pharmaceutical and agrochemical applications and potential to contribute to the understanding of the chemical ecology of bark beetle–microbe interactions in northern ecosystems.
1. Introduction
Small molecule natural products, or secondary/specialized metabolites, have historically been a significant source of active pharmaceutical agents [1,2,3]. Additionally, natural products are an emerging source for ecologically sustainable agrochemicals [4]. These secondary metabolites are produced by organisms in response to their environment, often unique to genera, species, and even bacterial strains [5]. Due to these constant ecological interactions, small molecule natural products are marked by their structural diversity and complex chemical scaffolds [6]. These structural scaffolds inspire synthetic advances, lead to molecular and biochemical understanding, and are the underpinnings of pharmaceuticals. New biologically relevant structural scaffolds drive chemical advances in these fields and thus the search for them. There is also an increased understanding that these secondary metabolites moderate microbiome interactions. As chemical diversification of these small molecule natural products (<2000 Daltons) occurs over evolutionary time, underexplored rich ecological niches can yield undiscovered natural products [7]. Northern sub-boreal and boreal forests are a diverse environmental resource whose natural product potential has yet to be significantly bioprospected.
Insect-associated microbes are often involved in complex ecological interactions which may be drivers for the production and diversification of specialized metabolites [8,9]. Indeed, it has been shown that insect-associated microbes are a rich source of novel natural products (NPs) with bioactive properties [10,11,12,13]. Canadian-isolated microbes currently represent only ~1% of natural product-relevant microbial databases, despite Canada’s biodiversity and size and despite the likelihood that unique ecological niches often yield novel chemistry [14]. The insect-associated bacterial natural product (NP) potential of northern sub-boreal and boreal forests [15] is of particular interest as it is largely unexplored. Endemic northern bark beetle (NBB) species are involved in complex tree–fungal–insect ecological relationships, important to forest dynamics in the Canadian provinces of British Columbia (BC), Alberta, and the Yukon Territory (YT) [16,17]. The fungal species help the bark beetles overcome tree host defenses and colonize bark galleries where the beetle larvae mature. Such complex insect relationships have previously been a rich source of bacterial NP chemodiversity and chemical ecology studies in tropical ants and termites [8,9,12,13]. Nevertheless, the natural product capacity of these NBB-associated bacteria is largely unexplored. Northern BC and Alberta have experienced pine (Dendroctonus ponderosae) and spruce (Dendroctonus rufipennis) beetle crises at varying times over the last 30 years, with their exploding populations causing devastation to both the forests and the forest industry. Two species, Dendroctonus ponderosae (mountain pine beetle—MPB) and D. rufipennis (spruce bark beetles—SBB), have been expanding far past their endemic range, causing devastation in the forests and forest industry. Recent research at the University of Northern British Columbia (UNBC) on the expanding D. ponderosae and D. rufipennis populations has shown strain level changes in the beetles and their fungal partners [18]. The chemical investigations of Streptomyces species isolated from the southern pine beetle (Dendroctonus frontalis) have shown that bacteria associated with Dendroctonus species can be a source of structural diversity and biologically relevant compounds [19]. Despite this, the chemical potential of northern Canadian pine and spruce beetles’ microbiomes or any of the endemic bark beetles has not been well investigated. In this work, we describe a first foray into exploring the natural product potential of bacteria isolated from a number of endemic NBB species from BC and YT in Canada utilizing biosynthetic gene cluster analysis and nitrogen-15 NMR.
Biosynthetic gene clusters (BGCs) are contiguous sets of genes within a microorganism’s genome that encode the enzymes and regulatory elements required for the production of secondary metabolites [20,21]. These natural products include antibiotics, antifungals, anticancer agents, and other bioactive compounds with ecological and therapeutic importance [22]. BGCs allow coordinated expression of genes for precursor biosynthesis, product assembly, and pre- and post-assembly tailoring.
Advances in genome sequencing and bioinformatics tools, such as antiSMASH, BiG-SCAPE, and MIBiG, have revolutionized the identification and analysis of BGCs [20,23,24]. These tools utilize core enzymes to predict the types of natural products encoded by BGCs—such as nonribosomal peptides (NRPs), polyketides (PKs), and ribosomally synthesized and post-translationally modified peptides (RiPPs)—and classify them based on sequence homology and domain architecture. Matching the chemical diversity of natural products, there is a corresponding diversity of BGCs. Structural differences in natural products can be hypothesized based on gene cluster organization differences and low sequence homology [25,26,27]. Thus, mining BGCs has become a powerful strategy for discovering novel bioactive compounds, offering a sustainable approach to drug discovery and development [21]. Nevertheless, this in silico prediction is just the first step, and natural products must still be isolated and structures elucidated in order for these compounds to be useful as pharmaceuticals and produced using chemical synthesis or biotechnological means [25,26,27].
Nuclear Magnetic Resonance (NMR) is a fundamental tool in natural product structure elucidation and discovery, capable of probing chemical environments of key elements, including nitrogen [28]. Among nitrogen’s isotopes, 15N is preferred for NMR studies due to its spin-½ properties and lack of quadrupolar relaxation, despite its low natural abundance (0.37%) and sensitivity [29]. Advances in two-dimensional experiments, such as 1H-15N HSQC and 1H-15N HSQC-TOCSY, have significantly enhanced the utility of nitrogen NMR by transferring sensitivity from 1H to 15N through polarization transfer [30,31]. This allows for the detection of nitrogen-containing functional groups in structurally complex and low-abundance natural products [30,31]. More recently, these advances have allowed 15N NMR to be utilized for targeted discovery of natural products in conjunction with genomic data [25,26,27,32,33,34].
Nitrogen functional groups are contained within nitrogen-containing natural products that have demonstrated bioactivities such as nonribosomal peptides (NRPs), nonribosomal peptide–polyketide hybrids (NRP-PK hybrids), alkaloids, and RiPPS. Nitrogen’s wide spectral range (~900 ppm) offers clear advantages in resolving chemical environments across diverse functional groups. Amides, amines, and nitrogen heterocycles exhibit distinct chemical shifts that can be directly linked to their electronic and structural environments [29,35]. For example: amides resonate between 150 ppm and 100 ppm; primary amines resonate between 80 and 30 ppm, influenced by hydrogen bonding and solvent effects; and nitrogen heterocycles, such as pyrroles and imidazoles, typically appear from 150 ppm to 250 ppm.
This wide spectral range is especially useful for nitrogen-containing secondary metabolites, such as peptides and alkaloids, where the large spectral dispersion of nitrogen facilitates observations of potential distinct natural product classes even within crude extracts. Recent methodological advances have made nitrogen NMR increasingly accessible for natural product analysis, even at low material quantities, by integrating high-field instruments and stable isotope labeling to enhance sensitivity [36]. This positions nitrogen NMR as an indispensable tool in identifying and characterizing nitrogen-rich metabolites from natural product libraries. Previous work has focused on utilizing nitrogen NMR in conjunction with gene cluster analysis for targeted discovery of specific natural product motifs.
In modern times, BGC data are beneficial, if not critical, for identifying bacterial natural product potential; however, many BGCs remain silent under standard laboratory conditions [37,38]. Previous studies have demonstrated that nitrogen-15 NMR experiments, like 1H-15N HSQC (detects directly bonded nitrogen protons), 1H-15N HSQC-TOCSY ( detects directly bonded nitrogen protons and associated spin systems), and 1H-15N HMBC (detects nitrogens and protons that are two–three bonds apart), can assess whether bacterial extracts contain expressed nitrogen-containing secondary metabolites [25,26,27,33,39]. The application of 15N NMR enables the detection of nitrogen-containing functional groups even in minimally processed crude extracts, allowing for experimental correlation with in silico natural product predictions [25,27]. The wide spectral dispersion of nitrogen NMR (~900 ppm) provides the resolution of functional groups such as amides, amines, and nitrogen heterocycles, which helps prioritize bacterial strains for isolation efforts targeting putatively novel nitrogen-containing natural products.
This study integrates BGC analysis with nitrogen-15 NMR to assess the chemical potential of bacteria associated with Canadian NBB. The primary objective is to establish an initial roadmap for the discovery of nitrogen-rich natural products by defining key screening criteria for prioritizing bacterial strains. Specifically, the study aims to identify bacterial isolates that contain (1) BGCs with low sequence homology or distinct gene organization and (2) correlations in ¹H-¹⁵N HSQC NMR experiments. Due to sequencing capacity constraints, bioassays are used as phenotypic guides to select bacterial strains from hundreds of isolates for whole genome sequencing. Once whole genome sequence data are obtained, bioinformatics tools such as antiSMASH and BiG-SCAPE are used to predict nitrogen-rich natural products, such as NRPs and NRP-PKS hybrids. By integrating genomic and ¹⁵N NMR spectroscopic approaches, this study seeks to determine whether predicted nitrogen-producing BGCs are actively expressed in initial isolate extract screens. These findings will guide the selection of bacterial strains for further media diversification, scale-up, and chemical investigations aimed at isolating and elucidating the structures of potentially novel nitrogen-containing scaffolds from northern Canada’s forests.
2. Materials and Methods
2.1. Sample Collection, Bacterial Isolation, and Cultivation
2.1.1. Sample Collection and Isolation
Samples were collected or donated from various bark beetle species and their galleries across BC and YT. Bark beetles were obtained from infected trees and funnel traps. Of the bacterial isolates discussed herein, collection sites were Whisker’s Point, BC, Canada (D. rufipennis and bark galleries); funnel traps laid by the Huber Lab at UNBC, Prince George, BC, Canada (D. pseudotsugae—Douglas Fir Beetles (DFB)); infected bolts provided by the BC Ministry of Forest from the Southern Kootenays (D. ponderosae); and funnel traps established and monitored by the YT Ministry of Forest in Haines Junction YT in Champagne–Aishik First Nations territory (Ips perturbatus). Before beetle collection, permission was granted by the Champagne–Aishik First Nations for retrieval of beetles from the funnel traps for scientific purposes, as well as a scientific permit received from the YT government. Beetle samples were processed immediately after collection or stored at −20 °C prior to processing.
Surface sterilization of the bark beetles was performed by submerging the beetles in a modified White’s solution (1 g HgCl2, 6.5 g NaCl, 1.25 L HCl, 250 mL ethanol, and 750 mL distilled water) for one minute [40]. Following sterilization via White’s solution, samples were rinsed with sterile ddH2O two times before further processing. This sterile water was spread on to TSA plates to ensure that no bacteria were present after sterilization. After sterilization, the bark beetle specimens were crushed in a sterile mortar and pestle, serially diluted with sterile water, and inoculated on selective media. Bark beetle frass collected from galleries of infected trees was placed into a sterile water and shaken for 24 h before being serially diluted onto selective media. Selective media included Actinomycete Isolation Agar (AIA), Burkholderia Isolation Media (RBMII), Chitin Agar II (CAII), Insect Frass Agar (IA), and Yeast Extract Malt Extract Agar (YEME) (see SI for complete media components). Plates were incubated at room temperature or 28 °C for up to 8 weeks, and distinct colonies were subcultured (min. 2×) onto further agar plates for purity and identification.
2.1.2. Cryogenic Storage of Bacterial Isolates
Pure bacterial isolates were cultured in Tryptic Soy Broth (TSB), MM1 broth, or MM11 broth and shaken (210 RPM) for 3–5 days at room temperature (Being Scientific Orbital Shaker, Canadawide Scientific, Ottawa, ON, Canada) or 28 °C (Thermofisher MaxQ 4000 Incubated Orbital Shaker, Fisher Scientific, Ottawa, ON, Canada). After growth, sterile 40% glycerol solution was added in a 1:1 ratio with the media containing the bacteria, mixed thoroughly, and dispensed in multiple sterile cryotubes. Thus, bacteria isolated were preserved in 20–25% glycerol for cryogenic storage at −80 °C (Thermofisher Cryogenic Freezer, Fisher Scientific, Ottawa, ON, Canada). All isolates were cataloged with unique identifiers in electronic and paper records.
2.1.3. Assays for Strain Prioritization and Subsequent Crude Extract Preparation
Pure bacterial isolates were screened for antifungal activity against Fusarium oxysporum and Fusarium culmorum. Challenge assays were conducted on YMS media (4 g yeast extract, 10 g malt extract, 4 g soluble starch, 20 g agar, and 1 L distilled water) as it was determined that the bacterial isolates and phytopathogens grew well on this media [41]. Protocols were performed as previously described [42]. The exception was the slow -growing Streptomyces bacterial cultures which were streaked on opposite poles of the plates, incubated for 4–5 days, and challenged with fungal plugs placed in the center of the plates. Zones of inhibition were categorized as strongly antifungal, weakly antifungal, or non-antifungal based on visible fungal suppression and previously described procedures [42].
As we must be selective in what strains we submit for whole genome sequencing due to lab resources, these initial assays were utilized to select bacterial strains with the potential for producing antifungal compounds to move forward with whole genome sequencing and spectroscopic analysis. After these assays were conducted, nine strains were selected (Table 1) for parallel analysis—(1) submission to sequencing labs for whole genome sequencing (Section 2.2) and (2) crude extract preparation to complete spectroscopic analysis and disc diffusion assays. Disc diffusion assays were conducted to check that the challenge assay results arose from organic solvent-soluble small molecule natural products, which are the compounds of interest for further study.

Table 1.
Sources and collection details of bacterial isolates from bark beetles and their galleries.
To begin crude extract preparation, 200 μl (100 mm plates) or 400 μl (150 mm) of bacterial inoculum was spread on agar plates; either three 100 mm plates or one 150 mm plate. Bacillus species were grown on RBMII, YMS, or PDA media. Streptomyces strains were grown on YMS or YEME agar for 14 days until sporulation was complete. All species were harvested by cutting the agar lawn into small squares, transferring to a glass container, and submerging with ethyl acetate (EtOAc, 100 mL) for extraction. After 24 h, the EtOAc extract was filtered through Whatman filter paper (Grade 2) and the agar resubmerged in fresh EtOAc (2×, 100 mL). All filtered EtOAc extracts were combined and the solvent removed under reduced pressured via rotary evaporation (Buchi R200) with a water bath held at 40 °C following similar protocols previously reported for crude extract preparation [25,26,27]. The resulting crude extract residues were dissolved in methanol (1–2 mL), transferred to pre-weighed 20 mL scintillation vials, and freeze dried (Buchi L-200) to remove residual solvents [25,41]. These crude extracts were stored in the freezer until ready to be examined in nitrogen NMR experiments and utilized for disc diffusion assays. No further filtrations steps were needed for NMR preparation, which is one of the strengths of NMR analysis of crude extracts.
For disc diffusion screens, crude extracts were reconstituted in DMSO at a concentration of 4 µg/µL for further assays. Disc diffusion assays were performed to evaluate the antifungal activity of crude extracts against Fusarium oxysporum. Crude extracts were pipetted onto Grade 3 Whatman filter paper discs, which were placed on YMS plates inoculated with fungal spore suspensions before being placed in a dark environment. Positive controls (10 mg/mL nystatin) and negative controls (DMSO) were included. Zones of inhibition were measured every 12 h beginning at 12 h after the initial incubation period and every 12 h thereafter until 72 h was reached.
2.2. Genomic Sequencing and BGC Analysis
Bacteria were grown for 24 h by shaking in tryptic soy broth at 28 °C or RT at 210 RPM (shakers listed in Section 2.1.2). After that growth, the cells were collected through centrifugation, the supernatant was discarded, and 500 uL of Zymoprep© DNA/RNA shield was added. Bacterial cells were then shipped to either EZbiome (KDM-22-177-SBBG, KDM-22-269-MPB, KDM-22-148-DFB, KDM-22-161-SBBG) or plasmidsaurus (KDM-22-185-DFB, KDM-23-16-IPS, KDM-23-410-IPS, KDM-23-158-IPS, KDM-23-275-IPS) for DNA extraction and whole genome sequencing. Short-read sequencing (EZbiome) was used for the KDM-22-177-SBBG, KDM-22-269-MPB, KDM-22-148-DFB, and KDM-22-161-SBBG strains, nanopore long-read sequencing (plasmidsaurus) was used for KDM-23-16-IPS, KDM-23-410-IPS, KDM-23-158-IPS, and KDM-23-275-IPS sequencing, and hybrid sequencing utilizing short reads and long reads from Illumina and nanopore sequencing systems, respectively, was utilized by plasmidsaurus for sequencing KDM-22-185-DFB. Sequencing data were obtained from the sequencing companies, and the FASTA files were edited utilizing Visual Studio Code v. 1.96 to add the lab strain names to the sequencing data. These files were then run through the bioinformatics software antiSMASH 7.0 both in the browser-based platform and natively on a downloaded antiSMASH [23]. The standard settings for the shell were modified to include ClusterBlast, MIBiG cluster comparison, and Cluster Pfam analysis [20,22]. After analysis, each antiSMASH result was reviewed at the gene cluster level, for known cluster homology, MIBiG comparison, and accuracy.
From the antiSMASH analysis, .gbk files for each predicted secondary metabolite gene cluster was downloaded and arranged into folders by bark beetle source, as well as a collective folder of all sequenced genomes for analysis with BiG-SCAPE-CORASON v.1 [24]. BiG-SCAPE-CORASON was installed using the Docker container provided by the developers through Git-Hub. Standard BiG-SCAPE settings were used to analyze the presence or absence of gene cluster families (GCFs) in bacteria that were sourced from the same bark beetles and across all bark beetles. Select natural product class networks obtained from BiG-SCAPE that demonstrated the presence of two or more gene clusters in the same GCF were visualized using Cytoscape v. 3.10.3 [43]. Select networks of interest, namely those natural product classes normally containing nitrogen, had their gene clusters further compared utilizing clinker [44].
2.3. NMR and Spectroscopic Profiling
1H-15N HSQC was chosen as the NMR experiment to probe the presence of nitrogen-containing natural products within crude extracts from select bacterial isolates as it has increased nitrogen sensitivity even from within unlabeled samples. To prepare samples for the nitrogen-15 NMR experiments, a minimum of 20 mg of crude extract was dissolved in 0.5 to 0.6 mL DMSO-d6 (Cambridge Isotopes, ACP Chemicals, Quebec, QC, Canada) and transferred to an appropriate NMR tube (5mm × 7 inches, Wilmad, Millipore Sigma, Toronto, ON, Canada). DMSO-d6 was selected as the solvent as it generally dissolves the greatest variety of polarities, and being non-protic, exchangeable protons on nitrogens are visible both in the 1H as well as 2D 1H-15N HSQC. All NMR experiments were run on a Bruker 400 MHz Evo NMR equipped with a SmartProbe. 1H-15N HSQC experiments were acquired with 256 scans in the direct dimension (F2) and 32 increments in the indirect dimension (F1). This greatly saves time but allows for increased sensitivity for observing unlabeled nitrogens with attached protons in the crude extracts. In our hands, long-range 1H-15N HMBC experiments are not sensitive enough to pick up nitrogen signals in crude extracts without labelling. F1 15N sweep width was set to 500 ppm with an O2P of 114 ppm.
2.4. General Experimental Procedures
The 15N NMR spectra were recorded on a Bruker Evo 400 MHz spectrometer equipped with a 2-channel multinuclear SmartProbe at 25 °C. 15N NMR two-dimensional spectra are referenced to ammonia set equal to 0 ppm. Chemical shift values have an accuracy of about 1 ppm. DMSO-d6 (99.9%), the deuterated solvent used for all NMR experiments listed in this paper, was purchased from Cambridge Isotopes through their Canadian distributor ACP Chemicals. All media components (Bioreagent grade, see SI for all detailed components) were obtained from Fisher Scientific or Millipore Sigma. All organic solvents were purchased from Millipore Sigma, Toronto, ON, Canada (EtOAc, ACS reagent grade; methanol, HPLC grade) or Fisher Scientific, Edmonton, Canada (DMSO, ACS grade). Zymoprep DNA/RNA shield was purchased through their Canadian distributor Cedarlane Labs (Canada).
2.5. List of Sample Name Abbreviations Used in the Text
- NBB—Northern bark beetle
- MPB—Mountain pine beetle (D. ponderosae)
- SBB—Spruce bark beetle (D. rufipennis)
- DFB—Douglas fir beetle (D. pseudotsugae)
- IPS—Ips perturbatus (a bark beetle species)
2.6. Use of Generative AI in Publication Preparation
During the preparation of this manuscript/study, the author(s) used Chat-GPT 4.0 to help assess written text for completion and flow. Text was written and then placed into Chat-GPT with the directives to ensure all listed data were complete as discussed in the text. It was also used to create table headings, to pull out a list of abbreviations from the completed text, and to correct grammar. At no time was Chat-GPT used for data creation or analysis. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
3. Results
3.1. Bacterial Isolation and Cultivation
Based on the initial challenge, pair-wise and disk diffusion assays, a total of nine bacterial isolates were selected from those obtained from bark beetles and their galleries, representing different species and collection sites in BC and YT (Table 1). Samples were collected predominantly from crushed beetles. MPB specimens were provided by Dr. Brent Murray (UNBC) collected from infected tree bolts. DFB specimens were provided by Dr. Dezene Huber (entomologist, UNBC) after collection from standard traps. Isolates from IPS were collected from crushed beetles obtained with the help of the YT Ministry of Forestry from standard traps they monitor near Haines Junction, YT. Two isolates were obtained from the bark beetle galleries left in the phloem of trees infected with SBB in Whisker’s Point, BC. The bacterial isolates were identified to species level using UBCG (Universal Bacterial Core Gene) analysis through Truebac ID vs 1 software [45]. Table 2 summarizes the closest bacterial species matches based on sequence identity and coverage. Most isolates were closely related to species within the genera Bacillus and Streptomyces. The highest sequence identity and coverage were observed for KDM-22-185-DFB, which matched Streptomyces cinereospinus at 99.4% identity and 94.8% coverage. Nevertheless, comparisons between the gene clusters of sequenced Streptomyces cinereospinus utilizing antiSMASH demonstrated little overlap in BGC identity. In contrast, KDM-22-161-SBBG exhibited lower identity and coverage values (91.0% and 42.6%, respectively), suggesting a more distant relationship to its closest match, Streptomyces brevispora.

Table 2.
Taxonomic identification of bacterial isolates based on sequence analysis.
3.2. Secondary Metabolite Biosynthetic Gene Cluster Analysis
3.2.1. Overview of Genomic Features
The genomes of nine bacterial isolates associated with bark beetles were sequenced and analyzed to determine genome size, number of contigs, GC content, and assembly quality metrics (N50). These features highlight the diversity and complexity of the isolates’ genomes (Table 3).

Table 3.
Genomic features of bacterial isolates from bark beetles.
Genome sizes ranged from 3.70 Mb (D. ponderosae, KDM-22-269-MPB) to 8.90 Mb (IPS, KDM-23-158-IPS), reflecting differences in genomic content across bacterial species. The number of contigs varied significantly from highly contiguous genomes with as few as 2 contigs (IPS, KDM-23-16-IPS and KDM-23-410-IPS) to more fragmented assemblies with up to 94 contigs (D. rufipennis, KDM-22-161-SBBG).
GC content was highly variable, with percentages ranging from 41.24% (D. ponderosae, KDM-22-269-MPB) to 73.46% (D. pseudotsugae, KDM-22-148-DFB). The high GC content observed in several isolates is typical for species within the Streptomyces genus. The Streptomyces genus is known to be a prolific producer of natural products and their high GC content. Indeed, it appears the Streptomyces isolates encode biosynthetic pathways for secondary metabolites, consistent with their predicted BGCs.
The assembly quality, measured by N50, varied widely, with the highest N50 value of 8,456,531 bp (IPS, KDM-23-16-IPS), indicating a highly contiguous assembly. Conversely, assemblies with smaller N50 values, such as 151,321 bp (D. pseudotsugae, KDM-22-148-DFB), were more fragmented, potentially due to genomic complexity or sequencing challenges.
3.2.2. BGC Identification and Analysis
BGC analysis was conducted from the genomes of nine bacterial isolates associated with bark beetles using antiSMASH 7.0, revealing a total of 226 BGCs across all samples (Table 4). These BGCs included 40 polyketide synthase (PKS) clusters, 29 nonribosomal peptide synthetase (NRPS) clusters, 23 PKS-NRPS hybrids, 31 terpene synthase clusters, and 43 RiPP clusters. A minimum of 42% of the BGCs were predicted to be coding for nitrogen-containing compounds—29 NRPS clusters, 23 PKS-NRPS hybrids, and 43 RiPP clusters, while not considering putative nitrogen-containing compounds among the “Other” category. Indeed, 60 gene clusters fell into the “Other” category, representing various less common biosynthetic pathways. antiSMASH automatically performs a comparison of predicted BGCs with the characterized BGCs from the MIBiG database through antiSMASH’s knownclusterblast and MIBiG comparison functions. By carefully assessing each predicted BGC against known clusters, it was observed that, notably, several NRPS and hybrid NRPS-PKS clusters exhibited unique domain organization and tailoring enzymes, suggesting the potential for novel bioactive compound production (see Figures S1–S3 for a few select examples).

Table 4.
Distribution of BGCs and their classes in bacterial isolates from bark beetles.
Cluster diversity across the isolates exhibited a wide range in the number and types of BGCs. KDM-22-177-SBBG, isolated from galleries of D. rufipennis-infected trees, contained 10 BGCs, predominantly PKS (five clusters), while KDM-22-161-SBBG harbored 30 BGCs, including seven PKS, seven NRPS, and two hybrid clusters. In isolates from D. pseudotsugae, KDM-22-185-DFB showed the highest diversity with 36 BGCs, including six hybrids and four terpene clusters. Among IPS isolates, KDM-23-16-IPS had the most BGCs (35), with 10 RiPP clusters, three hybrids, and four terpene clusters, while KDM-23-275-IPS exhibited similar diversity with 33 total BGCs and a balanced distribution across PKS, NRPS, hybrids, and other types.
These BGC values in the thirties aligns with previously reported Streptomyces quantities [46]. However, these genomes appear to have greater NRPS and PKS-NRPS BCG abundance (12% and 10%, respectively) compared to total values found in previous pangenome studies (5% and 2%) [46]. It is difficult to assess if this difference arises because the bacteria represented here were chosen for sequencing because of them producing a positive result in antifungal assays or if it is a result of their NBB association. Further genomic comparisons with NBB-associated bacteria that did not display such a phenotype in antifungal assays are needed to further assess any potential sources of differences.
Several genomes contained large numbers of partial or fragmented BGCs on contig edges. For example, KDM-22-161-SBBG had 15 partial fragments distributed across small contigs, likely representing one–three distinct BGCs. The total number of gene clusters for KDM-22-161-SBBG was assumed to have one additional BGC from the fragments. Similarly, KDM-22-148-DFB, KDM-22-177-SBBG, and KDM-23-275-IPS contained three, three, and one partial fragment, respectively, potentially belonging to additional BGCs or overlapping clusters that were not counted towards the total number of BGCs.
3.2.3. Gene Cluster Families, BGC Diversity, and Nitrogen-Containing BGCs
BGC analysis revealed that the Bacillus species, despite having fewer predicted gene clusters, overwhelmingly contained gene clusters with 90–100% homology to the BGCs of known compounds. In fact, multiple of these gene clusters (the majority of polyketide origin so not discussed herein) had 90–100% homology to antifungal compounds and are thus most likely producing the observed antifungal activity. Cblaster (CAGECAT) analysis revealed that the majority of these BGCs were common across other Bacillus species reported to have use as biopesticides or involved in microbiome interactions [47]. As such, these Bacillus species may play a role in the chemical ecology of the bark beetle–tree host–fungi–bacteria holobiont, but they are not promising reservoirs of novel natural products.
Conversely, analysis of the Streptomyces isolates revealed distinct gene clusters with low sequence homology (10–30%) to known BGCs. GCF analysis revealed that in the Streptomyces isolates, regardless of origin, BGCs for terpenes and RiPPS natural product classes were most commonly of high sequence homology to known compounds. In addition, predicted terpenes and RiPPS classes had the highest populated GCFs across isolates. Streptomyces albidoflavus strains KDM-22-148-DFB and KDM-23-410-IPS, like the Bacillus species, had BGCs with high sequence homology to known antifungal compounds of polyketide origin. Conversely, predicted NRPS and NPRS-PKS hybrids containing NRPS core domains in one or more modules appeared to have the most unique BGCs with little population of GCFs across the isolates. Clinker was thus applied to better visualize similarities [44].
Although not a nitrogen-containing product, terpenes were the most widely shared natural products between all the isolates. For the predicted terpene BGCs which contained at least one terpene synthase of the 31 identified terpene-like BGCs, BiG-SCAPE revealed that 24 were shared across multiple genomes, indicating a high degree of conservation among the isolates (Figure 1a). Notably, a terpene BGC associated with the terpene hopene was present in all Streptomyces genomes with 100% sequence homology and gene organization, while another terpene BGC with limited sequence homology to any characterized terpene synthase was identified in four of the sequenced genomes. This prevalence of hopene has been identified through pangenome analysis of Streptomyces species, as it has been shown to be the second most abundant BGC [46]. Given the role of terpene natural products in mediating bark beetle–tree host relationships, the identity of the unknown common terpenoid could be of interest to further chemical ecology studies [48].
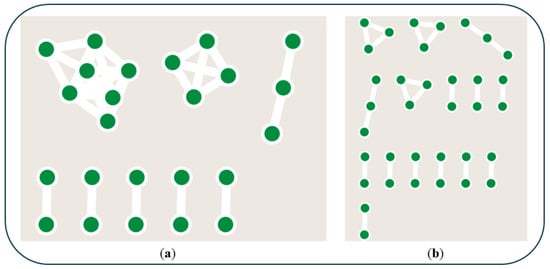
Figure 1.
(a) Terpene BGCs from the sequenced genomes grouped into families utilizing BiG-SCAPE and Cytoscape. (b) RiPP BGCs from the sequenced genomes grouped into families.
For the predicted RiPP BGCs, the 43 identified RiPP-like BGCs were predicted by BiG-SCAPE to exhibit substantial overlap, with 35 shared across at least two genomes (Figure 1b). This pattern suggests significant evolutionary conservation of RiPP biosynthesis within the sampled bacterial populations. Previous analysis of RiPP biosynthetic gene cluster distribution revealed that strains within the Actinobacterial clade tend to possess less potential (24%) for RiPP chemodiversity than those from the Proteobacteria phylum (70%) [49]. As the majority of strains in this study are in the Actinobacteria phylum, this result aligns with previous research. The NBB-associated bacteria do not appear to be a significant source of novel RiPP natural products.
Many nitrogen-containing natural products in bacteria belong to the nonribosomal peptide class and the nonribosomal peptide–polyketide hybrid structural class. These are produced by large multimodular NRPS and PKS-NRPS BGCs. The NRPS and PKS-NRPS hybrid BGCs were analyzed in detail using clinker to assess both whole-cluster similarity and diversity. Thirty-three of these BGCs were found to be distinct from one another, highlighting significant biosynthetic variability within the dataset (Figure 2 and Figure S1–S3). BiG-SCAPE-identified NRPS and NRPS-PKS hybrid BGC’s were visualized utilizing clinker. The clinker analysis determined a majority of unique BGCs with low sequence homology to characterized gene clusters. Among the 52 NRPS-like and PKS-NRPS hybrid BGCs, only one PKS-NRPS BGC—linked to the biosynthesis of [specific NRP families, e.g., frontalamide]—was shared across five genomes (Figure 3). An additional family, skyllamycin, was represented in three genomes (Figure 4). Thus, of the 52 predicted NRPS and NRPS-PKS BGCs, eight have high sequence homology to known natural products (15%) and 44 have low sequence homology to known natural products with characterized gene clusters (85%).
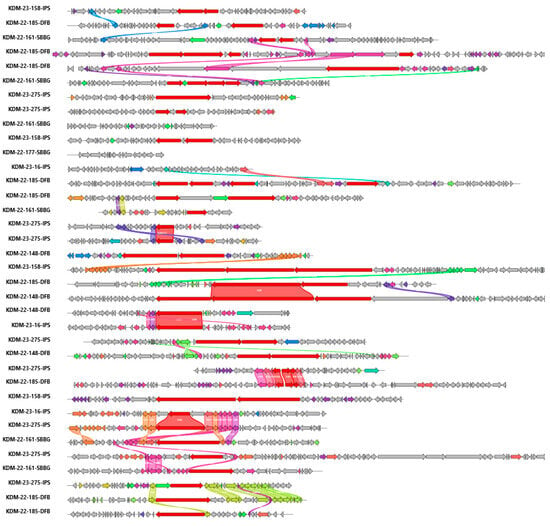
Figure 2.
Here, we see gene clusters across all nine NBB isolates containing NRPS modules or free domains with low sequence homology to characterized gene clusters. Gene cluster alignment and identity percentage was completed using clinker. Red-coloured genes indicate NRPS, all other colours randomly assigned by clinker to indicate closely related genes [44].
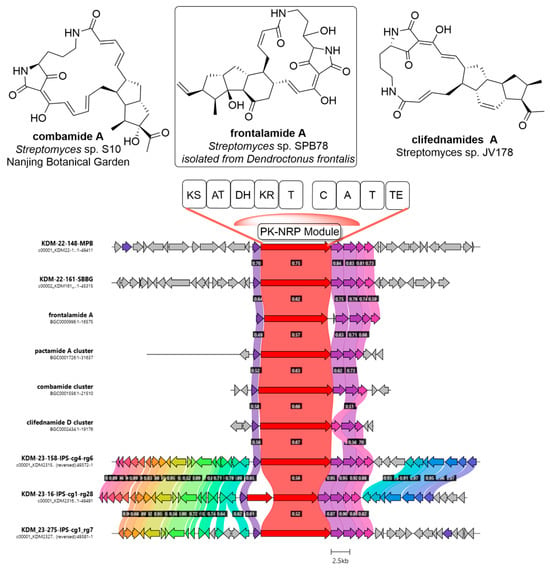
Figure 3.
Clinker analysis between five BGCs from across the nine isolates and characterized gene clusters in the frontalamide family. Red-coloured genes indicate NRPS, all other colours randomly assigned by clinker to indicate closely related genes. Gene cluster alignment and identity percentage was completed using clinker [44].
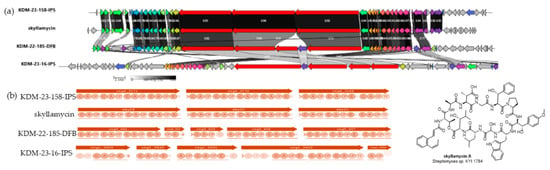
Figure 4.
Three BGCs across Streptomyces isolates were identified as bearing sequence homology to the nonribosomal peptide natural products family skyllamycins. (a) The gene cluster alignment between the BGCs from NBB-associated strains and the skyllamycin BGC was completed using clinker. Red-coloured genes indicate NRPS, all other colours randomly assigned by clinker to indicate closely related genes [44]. (b) Module and domain predictions of all BGCs retrieved from antiSMASH and MIBiG.
In isolates KDM-22-148-DFB, KDM-22-161-SBBG, KDM-23-158-IPS, KDM-23-16-IPS, and KDM-23-275-IPS, five NRPS BGCs appear to belong to an NRP-PKS hybrid family associated with the known polycyclic tetramate macrolactam family containing frontalamide, 10-epi-HSAF, SGR PTMs, pactamide A, montamide A, and combamide (Figure 3). Frontalamide was originally isolated from a southern bark beetle Dendroctonus frontalis [41]. Four bark beetle-associated isolates contained this BGC. This BGC has an unusual module that combines both NRP and PKS domains. The predicted frontalamide-encoding BGCs in the isolates contained the same domain organization as the characterized frontalamide gene cluster (Figure 3), save for KDM-23-16-IPS whose PKS module is predicted to be missing the dehydratase domain. This can be seen in the truncated module in Figure 3. Frontalamide is a polycyclic tetramate macrolactam which has demonstrated antifungal activity in the past. Due to this, it is possible that the demonstrated antifungal activity is associated with this natural product if it is being expressed. As strains of interest are investigated, dereplication strategies will be utilized to assess frontalamide’s production in various growth parameters. Moreover, due to the shared bark beetle origin of these bacteria, frontalamide may be an important natural product in bark beetle microbiome interactions. Given its previously demonstrated antifungal properties, testing pure frontalamide against NBB bark beetle fungal symbionts could provide valuable insights.
Additionally, isolates KDM-22-185-DFB, KDM-23-158-IPS, and KDM-23-16-IPS contained NRPS BGCs (Figure 4) that appear to belong to an NRP family associated with known compound skyllamycins. Skyllamycins are a class of heavily modified, nonribosomal depsipeptides with several structurally divergent members produced by unique Streptomyces strains isolated from the same New Zealand lichen Pseudocyphelleria dissimilis [50]. Specifically, KDM-22-185-DFB and KDM-23-158-IPS showed 100% similarity to known skyllamycin BGCs also observed in the domain organization, while KDM-23-16-IPS displayed only 35% similarity, indicating possible structural divergence or novel modifications. Analysis of the modules and domains of this BCG within KDM-23-16-IPS, demonstrates 8 modules rather than the 11 seen in skyllamycin along with domain variation. Skyllamycins have displayed antibacterial activity against Bacillus subtilis and to moderate cell growth by inhibiting platelet-derived growth factor, and so likely would not be involved in any of the antifungal activity we are seeing, if it were being expressed [50,51]. Furthermore, the majority of skyllamycins A, D, and E have been isolated from Streptomyces living in association with another living organism [50,51]. The presence of BGCs for these NRP in NBB-associated bacteria may indicate a role for these natural products in moderating ecological interactions. However, further studies along with isolation of the potentially novel variant would be required for a full investigation.
KDM-23-158-IPS and KDM-23-16-IPS demonstrated several GCFs containing BGCs with little homology to known compounds based on MIBiG and ClusterBlast comparisons (SI Figure S1). These findings underscore their potential as unexplored sources of new bioactive natural products.
Among the bacterial isolates, KDM-22-177-SBBG, KDM-22-269-MPB, and KDM-22-148-DFB harbored the highest proportion of known BGCs, with high homology to characterized compounds both containing nitrogen and not containing nitrogen. Conversely, isolates KDM-22-161-SBBG and KDM-22-185-DFB exhibited a higher proportion of orphan BGCs with no significant matches in the MIBiG or ClusterBlast databases, emphasizing their potential as sources of novel natural products.
3.3. Nitrogen-15 NMR and Spectroscopic Profiling
For the detection of nitrogen-containing compounds towards future isolation, analysis of crude extracts from selected bacterial isolates using 1H-15N HSQC NMR spectroscopy revealed the presence of nitrogen-containing metabolites, as indicated by distinct 1H-15N correlation signals. The number and chemical shift ranges of the observed signals varied across isolates, reflecting differences in their nitrogenous metabolite profiles (Table 5).

Table 5.
Detection of nitrogen-containing compounds in crude extracts via 1H-15N HSQC NMR.
KDM-22-161-SBBG, isolated from galleries of D. rufipennis-infected trees, exhibited the highest diversity of nitrogen signals, with 10 distinct correlations observed in the 1H-15N HSQC spectrum. Among these, eight signals were in the range of 100–150 ppm, characteristic of amide functionalities, aligning with the prediction of nine NRPS and NRP-PKS producing gene clusters in the bacterium’s genome. In addition, another was in the range > 150 ppm, likely corresponding to nitrogen heterocycles, and an additional signal was significantly more shielded. Neither of these correlations, however, bear direct analysis to predicted compound classes, although they are of interest and are currently being utilized to guide the isolation of nitrogen-containing compounds producing those chemical shifts in an active isolation effort.
For KDM-22-148-DFB, associated with Dendroctonus pseudotsugae, four nitrogen signals were detected, all within the range of 100–150 ppm, consistent with amides or other similar nitrogen-containing groups. This too aligns with the prediction of gene clusters producing nitrogen-containing compounds within this Streptomyces genome. Nevertheless, as mentioned, KDM-22-148-DFB contains known antifungals but of polyketides origin, and of its four predicted nitrogen-containing natural product BGCs, one demonstrates 75% sequence homology and identical gene organization to the BGC of the known compound frontalamide. As such, KDM-22-148-DFB is not of further interest for the discovery of novel compounds.
On the other hand, KDM-22-185-DFB, also associated with D. pseudotsugae, exhibited five nitrogen signals in its 1H-15N HSQC spectrum, with one additional signal appearing in the <100 ppm range. This distribution suggests the presence of both amide functionalities and primary or secondary amines. Notably, genome analysis revealed the presence of several NRPS and hybrid NRPS-PKS gene clusters, aligning with these spectroscopic observations. Additionally, KDM-22-185-DFB harbors a biosynthetic gene cluster related to the production of azinothricin (SI Figure S3), a structurally complex nitrogen-containing natural product with potential antimicrobial properties. The presence of an unusual gene organization within this cluster suggests possible structural divergence from known azinothricins. In addition, the shielded nitrogen signal <100 ppm correlates with a proton signal at 5.5 ppm in the 1H-15N HSQC experiment (SI Figure S10). The appearance of this signal could correlate with the expression of an azinothricin natural product as these signals of a shielded nitrogen and relatively shielded proton signal have been used for the discovery of piperazic acid-containing natural products in prior discovery efforts. Thus, the combination of BGC analysis and nitrogen-15 NMR application has reinforced the potential for novel nitrogen-rich natural products, and extracts from this strain are currently under active isolation efforts.
The Ips perterbatus-associated isolates displayed varying nitrogen signal profiles:
KDM-23-16-IPS showed three signals, with one in the <100 ppm range (possibly primary or secondary amines) and two in the 100–150 ppm range. This aligns with the fewer predicted BGCs for NRP and NRP-PKS (5).
KDM-23-410-IPS exhibited only one nitrogen signal in the 100–150 ppm range, while KDM-23-158-IPS had three signals exclusively in this range. KDM-23-275-IPS showed a single signal in the <100 ppm range, indicative of amine groups. The predicted NRP and NRP-PKS BGCs in these three strains were more abundant than displayed in the 1H-15N HSQC NMR. This suggests that the media growth conditions may not have triggered the production of the predicted natural products. Further investigation would require growth in alternative media and analysis using 1H-15N HSQC NMR.
For the Bacillus species, KDM-22-177-SBBG or KDM-22-269-MPB, the only nitrogen signals were detected below 100 ppm, suggesting few detectable nitrogen-rich metabolites in the media grown or concentrations below the detection threshold. These species do have smaller genomes and few predicted NRP and NRP-PKS BGCs.
These findings highlight the diversity of nitrogenous metabolites potentially produced by NBB-associated bacteria and underscore the utility of 1H-15N HSQC in profiling crude extracts for prioritization of bacterial strains for the intense efforts needed for the isolation and structure elucidation of novel nitrogen-containing compounds. The presence of signals across a wide spectral range suggests the potential for varied bioactive compounds, including amides, amines, and nitrogen heterocycles. Further isolation and structural elucidation are needed to characterize these natural products.
The exploration of nitrogen-containing natural products through the combined use of BGC analysis and nitrogen-15 NMR spectroscopy has provided valuable insights into the chemical potential of northern Canadian bark beetle-associated bacteria. The detection of nitrogen-rich metabolites in crude extracts and the diversity of their predicted biosynthetic pathways highlight the utility of this integrated approach in uncovering novel bioactive compounds.
This study represents the first BGC analysis of bacteria associated with diverse endemic Canadian NBB. The combined application of genomic and nitrogen-15 NMR spectroscopy has provided valuable insights into the chemical potential of these bacteria, particularly in the discovery of nitrogen-containing natural products. BGC analysis using BiG-SCAPE and clinker revealed extensive diversity among the isolates, particularly in nitrogen-associated NRPS, NRPS-PKS hybrids, and RiPP clusters. RiPP clusters demonstrated a significant overlap of BGCs between unique Streptomyces species with no singletons in the BiG-SCAPE analysis with 43 predicted BGCs in fourteen GCFs. The NRPS and NRPS-PKS hybrids demonstrated a greater amount of BGCs with low sequence homology either to characterized BGCs (via MIBiG analysis) or to other NBB-associated Streptomyces (via BiG-SCAPE and clinker). Of the 52 NRP- and NRP-PKS-predicted BGCs, eight demonstrated high sequence homology (>50%) and identical or near identical gene organization to characterized gene clusters, whereas three BGCs belonged to an NBB-associated GCF with low sequence homology to characterized gene clusters. The rest of these BGCs (38% of all RiPP, NRP, and NRP-PKS) presented as singletons in both BiG-SCAPE and clinker and with low sequence homology or unusual gene organization when compared to characterized gene clusters. This level of unique BGCs (~40%) is consistent with other Actinomycete studies from extreme environments, such as polar regions [52]. Streptomyces species consistently harbored more BGCs than Bacillus strains, which is expected given the well-established role of Streptomyces as prolific producers of bioactive secondary metabolites [53].
In comparison to another BGC analysis of Streptomyces strains, NBB-associated Streptomyces showed similar numbers of BGCs per genome (around 30–36) [46]. However, select NBB-associated Streptomyces isolates exhibited an increased proportion of BGCs coding for nitrogen-containing metabolites in comparison to pangenome studies, suggesting that these bacteria may be a particularly rich source of nitrogenous natural products [46]. The reason for this increased nitrogen–BGC ratio remains an open question—whether it arises due to the antifungal selection strategy prior to whole genome sequencing or whether it is a feature of NBB association requires further study. Despite the presence of some known BGCs, the gene cluster analysis provided a rapid means to deprioritize strains that predominantly produce previously characterized compounds while highlighting at least four strains with strong potential for novel nitrogen-containing chemistry. Genome mining in different capacities has been utilized for natural product discovery, particularly microbial natural product discovery, and these findings continue to support the strengths of genome mining and the utility of different tools that have been developed in the field for BGC analysis [21,37].
The integration of nitrogen-15 NMR spectroscopy further strengthened the prioritization of these isolates. As a non-destructive screening method, nitrogen-15 NMR enables the direct detection of nitrogenous functional groups in crude extracts with minimal sample processing. The 1H-15N HSQC and 1H-15N HMBC (with enough material or stable isotope labelling) experiments only detect nitrogen-containing natural products, so data analysis is streamlined. Pairing genomic methods with nitrogen-15 NMR allows prioritization of those strains that possess BGCs of interest and are producing nitrogen-containing natural products under laboratory conditions. Future isolation efforts are prioritizing strains with unique BGC and NMR profiles, particularly those exhibiting nitrogen signals in underrepresented spectral regions.
Notably, for some isolates, the number of nitrogen signals observed in the 1H-15N HSQC spectra was lower than the number of BGCs that could putatively produce nitrogen-containing natural products. This suggests that media growth conditions may not have induced the expression of all biosynthetic pathways, highlighting the need for media diversification studies. 1H-15N HSQC will provide a fast and robust method for exploring alternative culture conditions that could enhance the production of nitrogenous metabolites. A key advantage of nitrogen-15 NMR screening is its minimal sample preparation requirements—crude extracts require little processing before NMR analysis. Furthermore, after NMR analysis, DMSO-d6 can be removed from the crude extract without any loss in material, permitting the application of other techniques such as mass spectrometry, bioassays, or separation methods.
Building on recent advancements in NMR-based metabologenomic strategies, this study highlights the power of integrating genomic predictions with nitrogen-15 NMR spectroscopy for strain prioritization [25,27,32,33,39,54]. By combining these approaches, researchers can efficiently identify the bacterial strains most likely to yield novel nitrogen-containing bioactive compounds, laying the groundwork for future isolation, structural characterization, and functional studies. From that efficient isolation, novel structures can expand our understanding of the chemical ecology of NBB-associated bacteria and also provide nitrogen-rich natural products with potential pharmaceutical and agrochemical applications.
4. Conclusions
This study provides the first analysis of BGCs for nitrogen-containing natural products from bacteria associated with northern Canadian bark beetles. Through the application of multiple secondary metabolite bioinformatic software platforms including antiSMASH for BGC identification, MIBiG for the comparison of predicted BGCs with characterized gene clusters, BiG-SCAPE for the identification of GCFs, and clinker for the visualization and identification of GCFs, NBB-associated bacteria were identified to harbor a diverse array of BGCs. This was particularly observed with the nitrogen-associated NRPS and NRPS-PKS hybrid BGCs. While RiPP clusters exhibited substantial conservation across strains, NRPS and NRPS-PKS hybrids demonstrated high diversity, with a significant proportion showing low sequence homology to known biosynthetic pathways. These findings suggest that bark beetle-associated bacteria represent an underexplored resource for novel nitrogen-containing secondary metabolites.
By integrating nitrogen-15 NMR spectroscopy with genomic analysis, we have established a robust workflow for prioritizing strains that actively produce nitrogen-containing compounds under laboratory conditions. The rapid and non-destructive nature of nitrogen-15 NMR allowed for efficient screening of crude extracts, providing direct evidence of predicted nitrogenous secondary metabolites being expressed under laboratory conditions. This approach effectively streamlined the identification of high-priority strains for future targeted isolation and structural characterization.
The broader impact of this work extends beyond natural product discovery. The identification of nitrogen-containing natural products from bark beetle-associated bacteria has implications for biocontrol strategies, microbial ecology, and the development of novel antifungal, antibacterial, and agrochemical agents. This study underscores the value of coupling genomic mining with spectroscopic tools in microbial natural product discovery and sets a foundation for future exploration of Canada’s boreal and sub-boreal microbiomes as reservoirs of bioactive compounds with potential pharmaceutical and agricultural applications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/nitrogen6010013/s1. Additional BGC data and NMR Spectroscopy data can be found in the supplementary materials.
Author Contributions
Conceptualization, K.D.M., methodology, N.A., K.D.M., N.J., H.S., M.B. and M.S.; software, K.D.M.; validation, K.D.M.; formal analysis, K.D.M.; investigation, N.A., K.D.M., N.J., H.S., M.B. and M.S.; resources, K.D.M.; data curation, K.D.M.; writing—original draft preparation, K.D.M., N.A., N.J., H.S. and M.S.; writing—review and editing, K.D.M.; visualization, K.D.M.; supervision, K.D.M.; project administration, K.D.M. and N.J.; funding acquisition, K.D.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NSERC Discovery Grant, grant number RGPIN-2023-05625 and UNBC start-up funds (no grant number).
Data Availability Statement
The original data presented in the study are openly available at the National Centre for Biotechnology Information (NCBI) for the strains KDM-22-177-SBBG, KDM-22-269-MPB, KDM-22-148-DFB, and KDM-23-275-IPS, which will not be investigated further for novel natural products. These whole genome sequencing data for these strains have been deposited at NCBI with the BioSample accessions: SAMN46853761, SAMN46853762, SAMN46853763, and SAMN46853764 Release date: 2026-03-31. The remaining whole genome sequencing datasets presented in this article are not readily available because the datasets are part of an ongoing study.
Acknowledgments
The authors gratefully acknowledge Brent Murray and Dezene Huber (UNBC) for their assistance in obtaining and identifying bark beetle species and for their guidance on the use of funnel traps.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, S.; Tiezzi, A.; Laghezza Masci, V.; Ovidi, E. Natural Products for Human Health: An Historical Overview of the Drug Discovery Approaches. Nat. Prod. Res. 2018, 32, 1926–1950. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Natural Products Discovery and Potential for New Antibiotics. Curr. Opin. Microbiol. 2019, 51, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Gerwick, B.C.; Sparks, T.C. Natural Products for Pest Control: An Analysis of Their Role, Value and Future. Pest Manag. Sci. 2014, 70, 1169–1185. [Google Scholar] [CrossRef] [PubMed]
- Choudoir, M.J.; Pepe-Ranney, C.; Buckley, D.H. Diversification of Secondary Metabolite Biosynthetic Gene Clusters Coincides with Lineage Divergence in Streptomyces. Antibiotics 2018, 7, 12. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Park, C.J.; Caimi, N.A.; Buecher, D.C.; Valdez, E.W.; Northup, D.E.; Andam, C.P. Unexpected Genomic, Biosynthetic and Species Diversity of Streptomyces Bacteria from Bats in Arizona and New Mexico, USA. BMC Genom. 2021, 22, 247. [Google Scholar] [CrossRef]
- Beemelmanns, C.; Guo, H.; Rischer, M.; Poulsen, M. Natural Products from Microbes Associated with Insects. Beilstein J. Org. Chem. 2016, 12, 314–327. [Google Scholar] [CrossRef]
- Chanson, A.; Moreau, C.S.; Duplais, C. Assessing Biosynthetic Gene Cluster Diversity of Specialized Metabolites in the Conserved Gut Symbionts of Herbivorous Turtle Ants. Front. Microbiol. 2021, 12, 1640. [Google Scholar] [CrossRef]
- Arnam, E.B.V.; Currie, C.R.; Clardy, J. Defense Contracts: Molecular Protection in Insect-Microbe Symbioses. Chem. Soc. Rev. 2018, 47, 1638–1651. [Google Scholar] [CrossRef]
- Oh, D.-C.; Poulsen, M.; Currie, C.R.; Clardy, J. Dentigerumycin: A Bacterial Mediator of an Ant-Fungus Symbiosis. Nat. Chem. Biol. 2009, 5, 391–393. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Carlson, C.M.; Ortega, H.E.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The Antimicrobial Potential of Streptomyces from Insect Microbiomes. Nat. Commun. 2019, 10, 516. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Ko, H.; Bang, H.-S.; Park, S.-H.; Kim, D.-G.; Kwon, H.C.; Kim, S.Y.; Shin, J.; Oh, D.-C. Coprismycins A and B, Neuroprotective Phenylpyridines from the Dung Beetle-Associated Bacterium, Streptomyces sp. Bioorganic Med. Chem. Lett. 2011, 21, 5715–5718. [Google Scholar] [CrossRef] [PubMed]
- Dalisay, D.S.; Williams, D.E.; Wang, X.L.; Centko, R.; Chen, J.; Andersen, R.J. Marine Sediment-Derived Streptomyces Bacteria from British Columbia, Canada Are a Promising Microbiota Resource for the Discovery of Antimicrobial Natural Products. PLoS ONE 2013, 8, e77078. [Google Scholar] [CrossRef] [PubMed]
- Six, D.L. The Bark Beetle Holobiont: Why Microbes Matter. J. Chem. Ecol. 2013, 39, 989–1002. [Google Scholar] [CrossRef]
- Sickle, A.V.; Fiddick, R.L.; Wood, C.S. History of Forest Insect Investigations in British Columbia II. The Forest Insect and Disease Survey in the Pacific Region. J. Entomol. Soc. Br. Columbia 2001, 98, 169–176. [Google Scholar]
- 2021 Yukon Forest Health Report 2022. Available online: https://yukon.ca/en/2022-yukon-forest-health-report (accessed on 1 June 2023).
- Gayathri Samarasekera, G.D.N.; Bartell, N.V.; Lindgren, B.S.; Cooke, J.E.K.; Davis, C.S.; James, P.M.A.; Coltman, D.W.; Mock, K.E.; Murray, B.W. Spatial Genetic Structure of the Mountain Pine Beetle (Dendroctonus ponderosae) Outbreak in Western Canada: Historical Patterns and Contemporary Dispersal. Mol. Ecol. 2012, 21, 2931–2948. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-C.; Scott, J.J.; Currie, C.R.; Clardy, J. Mycangimycin, a Polyene Peroxide from a Mutualist Streptomyces sp. Org. Lett. 2009, 11, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Muñoz, J.C.; Terlouw, B.R.; van der Hooft, J.J.J.; van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V.; et al. MIBiG 2.0: A Repository for Biosynthetic Gene Clusters of Known Function. Nucleic Acids Res. 2020, 48, D454–D458. [Google Scholar] [CrossRef] [PubMed]
- Chevrette, M.G.; Gavrilidou, A.; Mantri, S.; Selem-Mojica, N.; Ziemert, N.; Barona-Gómez, F. The Confluence of Big Data and Evolutionary Genome Mining for the Discovery of Natural Products. Nat. Prod. Rep. 2021, 38, 2024–2040. [Google Scholar] [CrossRef] [PubMed]
- Zdouc, M.M.; Blin, K.; Louwen, N.L.L.; Navarro, J.; Loureiro, C.; Bader, C.D.; Bailey, C.B.; Barra, L.; Booth, T.J.; Bozhüyük, K.A.J.; et al. MIBiG 4.0: Advancing Biosynthetic Gene Cluster Curation through Global Collaboration. Nucleic Acids Res. 2025, 53, D678–D690. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Morgan, K.D.; Williams, D.E.; Patrick, B.O.; Remigy, M.; Banuelos, C.A.; Sadar, M.D.; Ryan, K.S.; Andersen, R.J. Incarnatapeptins A and B, Nonribosomal Peptides Discovered Using Genome Mining and 1H/15N HSQC-TOCSY. Org. Lett. 2020, 22, 4053–4057. [Google Scholar] [CrossRef] [PubMed]
- Morgan, K.D.; Williams, D.E.; Ryan, K.S.; Andersen, R.J. Dentigerumycin F and G: Dynamic Structures Retrieved through a Genome-Mining/Nitrogen-NMR Methodology. Tetrahedron Lett. 2022, 94, 153688. [Google Scholar] [CrossRef]
- Hagar, M.; Morgan, K.D.; Stumpf, S.D.; Tsingos, M.; Banuelos, C.A.; Sadar, M.D.; Blodgett, J.A.V.; Andersen, R.J.; Ryan, K.S. Piperazate-Guided Isolation of Caveamides A and B, Cyclohexenylalanine-Containing Nonribosomal Peptides from a Cave Actinomycete. Org. Lett. 2024, 26, 4127–4131. [Google Scholar] [CrossRef]
- Breton, R.C.; Reynolds, W.F. Using NMR to Identify and Characterize Natural Products. Nat. Prod. Rep. 2013, 30, 501. [Google Scholar] [CrossRef]
- Witanowski, M.; Webb, G.A. Nitrogen NMR Spectroscopy. In Annual Reports on NMR Spectroscopy; Mooney, E.F., Ed.; Academic Press: London, UK, 1972; Volume 5, pp. 395–464. ISBN 978-0-12-505305-1. [Google Scholar]
- Martin, G.E.; Williams, A.J. Utilizing Long-Range 1 H–15 N 2-D NMR Spectroscopy for Chemical Structure Elucidation and Confirmation. In eMagRes; Harris, R.K., Wasylishen, R.L., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2010; ISBN 978-0-470-03459-0. [Google Scholar]
- Marek, R.; Lycka, A. 15N NMR Spectroscopy in Structural Analysis. COC 2002, 6, 35–66. [Google Scholar] [CrossRef]
- Hu, J.-Q.; Wang, J.-J.; Li, Y.-L.; Zhuo, L.; Zhang, A.; Sui, H.-Y.; Li, X.-J.; Shen, T.; Yin, Y.; Wu, Z.-H.; et al. Combining NMR-Based Metabolic Profiling and Genome Mining for the Accelerated Discovery of Archangiumide, an Allenic Macrolide from the Myxobacterium Archangium violaceum SDU8. Org. Lett. 2021, 23, 2114–2119. [Google Scholar] [CrossRef]
- Shin, D.; Byun, W.S.; Kang, S.; Kang, I.; Bae, E.S.; An, J.S.; Im, J.H.; Park, J.; Kim, E.; Ko, K.; et al. Targeted and Logical Discovery of Piperazic Acid-Bearing Natural Products Based on Genomic and Spectroscopic Signatures. J. Am. Chem. Soc. 2023, 145, 19676–19690. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Reheman, A.; Wan, C. Discovery of Anti-Mycobacterium Tuberculosis Desertomycins from Streptomyces Flavofungini TRM90047 Based on Genome Mining and HSQC-TOCSY. Sci. Rep. 2024, 14, 17006. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.T.; Márquez, B.L.; Gerwick, W.H. Use of 1H-15N PEP-HSQC-TOCSY at Natural Abundance to Facilitate the Structure Elucidation of Naturally Occurring Peptides. Tetrahedron 1999, 55, 2881–2888. [Google Scholar] [CrossRef]
- Morgan, K.D. The Use of Nitrogen-15 in Microbial Natural Product Discovery and Biosynthetic Characterization. Front. Microbiol. 2023, 14, 1174591. [Google Scholar] [CrossRef]
- Bauman, K.D.; Butler, K.S.; Moore, B.S.; Jonathan, R. Chekan Genome Mining Methods to Discover Bioactive Natural Products. Nat. Prod. Rep. 2021. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.-H.; Im, J.H.; Kang, I.; Kim, E.; Jang, S.C.; Cho, E.; Shin, D.; Hwang, S.; Du, Y.E.; Huynh, T.-H.; et al. Genomic and Spectroscopic Signature-Based Discovery of Natural Macrolactams. J. Am. Chem. Soc. 2023, 145, 1886–1896. [Google Scholar] [CrossRef]
- Barras, S.J. Improved White’s Solution for Surface Sterilization of Pupae of Dendroctonus Frontalis1. J. Econ. Entomol. 1972, 65, 1504. [Google Scholar] [CrossRef] [PubMed]
- Blodgett, J.A.V.; Oh, D.-C.; Cao, S.; Currie, C.R.; Kolter, R.; Clardy, J. Common Biosynthetic Origins for Polycyclic Tetramate Macrolactams from Phylogenetically Diverse Bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11692–11697. [Google Scholar] [CrossRef]
- Lauer, A.; Baal, J.D.; Mendes, S.D.; Casimiro, K.N.; Passaglia, A.K.; Valenzuela, A.H.; Guibert, G. Valley Fever on the Rise—Searching for Microbial Antagonists to the Fungal Pathogen Coccidioides Immitis. Microorganisms 2019, 7, 31. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap.Js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.-M.; Kim, C.K.; Roh, J.; Byun, J.-H.; Yang, S.-J.; Choi, S.-B.; Chun, J.; Yong, D. Application of the Whole Genome-Based Bacterial Identification System, TrueBac ID, Using Clinical Isolates That Were Not Identified With Three Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) Systems. Ann. Lab. Med. 2019, 39, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Mohite, O.S.; Jørgensen, T.S.; Booth, T.J.; Charusanti, P.; Phaneuf, P.V.; Weber, T.; Palsson, B.O. Pangenome Mining of the Streptomyces Genus Redefines Species’ Biosynthetic Potential. Genome Biol. 2025, 26, 9. [Google Scholar] [CrossRef] [PubMed]
- Van Den Belt, M.; Gilchrist, C.; Booth, T.J.; Chooi, Y.-H.; Medema, M.H.; Alanjary, M. CAGECAT: The CompArative GEne Cluster Analysis Toolbox for Rapid Search and Visualisation of Homologous Gene Clusters. BMC Bioinform. 2023, 24, 181. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.S.; Boone, C.K.; Bohlmann, J.; Raffa, K.F. Responses of Bark Beetle-Associated Bacteria to Host Monoterpenes and Their Relationship to Insect Life Histories. J. Chem. Ecol. 2011, 37, 808–817. [Google Scholar] [CrossRef]
- Malit, J.J.L.; Wu, C.; Liu, L.-L.; Qian, P.-Y. Global Genome Mining Reveals the Distribution of Diverse Thioamidated RiPP Biosynthesis Gene Clusters. Front. Microbiol. 2021, 12, 635389. [Google Scholar] [CrossRef] [PubMed]
- Bracegirdle, J.; Hou, P.; Nowak, V.V.; Ackerley, D.F.; Keyzers, R.A.; Owen, J.G. Skyllamycins D and E, Non-Ribosomal Cyclic Depsipeptides from Lichen-Sourced Streptomyces Anulatus. J. Nat. Prod. 2021, 84, 2536–2543. Available online: https://pubs-acs-org.prxy.lib.unbc.ca/doi/10.1021/acs.jnatprod.1c00547 (accessed on 16 February 2025). [CrossRef]
- Toki, S.; Agatsuma, T.; Ochiai, K.; Saitoh, Y.; Ando, K.; Nakanishi, S.; Lokker, N.A.; Giese, N.A.; Matsuda, Y. RP-1776, a Novel Cyclic Peptide Produced by Streptomyces sp., Inhibits the Binding of PDGF to the Extracellular Domain of Its Receptor. J. Antibiot. 2001, 54, 405–414. [Google Scholar] [CrossRef]
- Soldatou, S.; Eldjárn, G.H.; Ramsay, A.; van der Hooft, J.J.J.; Hughes, A.H.; Rogers, S.; Duncan, K.R. Comparative Metabologenomics Analysis of Polar Actinomycetes. Mar. Drugs 2021, 19, 103. [Google Scholar] [CrossRef]
- Baltz, R.H. Gifted Microbes for Genome Mining and Natural Product Discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef]
- Xu, S.; Wang, N.; Meng, Q.; Ma, W.; Li, H. Metabologenomics-Driven Discovery of Nocardimicins from a Psychrophilic Nocardia Sp. Strain. J. Nat. Prod. 2025, 88, 103–109. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).