Do the Leaves of Multiple Invasive Plants Decompose More Easily than a Native Plant’s under Nitrogen Deposition with Different Forms?
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Invasive Plants
2.2. Selection of Native Plant
2.3. Experimental Design
2.4. Determination of the Decomposition Rate
2.5. Determination of Soil Physicochemical Properties and Enzyme Activities
2.6. Determination of the Structure of Soil Bacterial Community
2.7. Statistical Analysis
3. Results
3.1. Differences in Decomposition Rate
3.2. Differences in Soil Physicochemical Properties and Enzyme Activities
3.3. Differences in Soil Bacterial Alpha Diversity
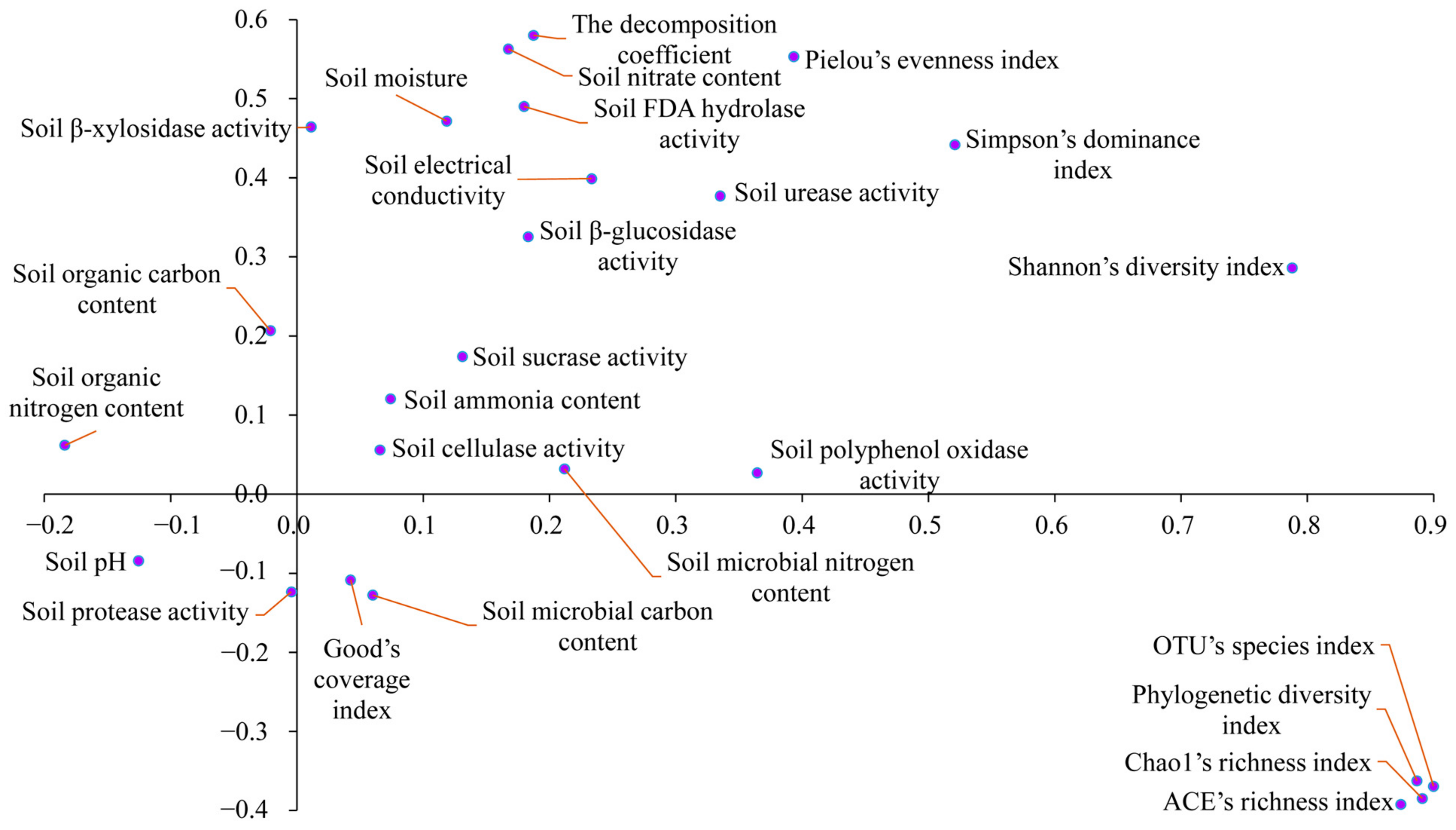
3.4. Correlation Patterns between Soil Physicochemical Properties and Enzyme Activities, SBAD, and Decomposition Coefficient
3.5. Differences in the Structure of Soil Bacterial Community
3.6. Differences in the Number of Functional Gene Pathways of Soil Bacterial Community Involved in the Decomposition Process under D-AN with Four Forms
3.7. Differences in the Number of Functional Gene Pathways of Soil Bacterial Community Involved in the Decomposition Process Treated with the Leaves of the Four AIPs and P. laciniata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forey, E.; Lodhar, S.Y.F.; Galvin, S.D.; Lowry, J.H.; Gopaul, S.; Hanson, G.; Carboni, M.; Chauvat, M.; Boehmer, H.J. Alien palm invasion leads to selective biotic filtering of resident plant communities towards competitive functional traits. Biol. Invasions 2023, 25, 1489–1508. [Google Scholar] [CrossRef]
- Beshai, R.A.; Truong, D.A.; Henry, A.K.; Sorte, C.J.B. Biotic resistance or invasional meltdown? Diversity reduces invasibility but not exotic dominance in southern California epibenthic communities. Biol. Invasions 2023, 25, 533–549. [Google Scholar] [CrossRef]
- Erckie, L.; Adedoja, O.; Geerts, S.; van Wyk, E.; Boatwright, J.S. Impacts of an invasive alien Proteaceae on native plant species richness and vegetation structure. S. Afr. J. Bot. 2022, 144, 332–338. [Google Scholar] [CrossRef]
- Czortek, P.; Królak, E.; Borkowska, L.; Bielecka, A. Effects of surrounding landscape on the performance of Solidago canadensis L. and plant functional diversity on heavily invaded post-agricultural wastelands. Biol. Invasions 2023, 25, 2477–2494. [Google Scholar] [CrossRef]
- Yan, X.L.; Liu, Q.R.; Shou, H.Y.; Zeng, X.F.; Zhang, Y.; Chen, L.; Liu, Y.; Ma, H.Y.; Qi, S.Y.; Ma, J.S. The categorization and analysis on the geographic distribution patterns of Chinese alien invasive plants. Biodivers. Sci. 2014, 22, 667–676. [Google Scholar]
- Wang, C.Y.; Liu, J.; Xiao, H.G.; Zhou, J.W.; Du, D.L. Floristic characteristics of alien invasive seed plant species in China. An. Acad. Bras. Cienc. 2016, 88, 1791–1797. [Google Scholar] [CrossRef] [PubMed]
- Ahmad Dar, M.; Ahmad, M.; Singh, R.; Kumar Kohli, R.; Singh, H.P.; Batish, D.R. Invasive plants alter soil properties and nutrient dynamics: A case study of Anthemis cotula invasion in Kashmir Himalaya. Catena 2023, 226, 107069. [Google Scholar] [CrossRef]
- Kone, A.W.; Kassi, S.P.A.Y.; Koffi, B.Y.; Masse, D.; Maiga, A.A.; Tondoh, J.E.; Kisaka, O.M.; Toure, G.P.T. Chromolaena odorata (L.) K&R (Asteraceae) invasion effects on soil microbial biomass and activities in a forest-savanna mosaic. Catena 2021, 207, 105619. [Google Scholar] [CrossRef]
- Cerrato, M.D.; Cortés-Fernández, I.; Ribas-Serra, A.; Mir-Rosselló, P.M.; Cardona, C.; Gil, L. Time pattern variation of alien plant introductions in an insular biodiversity hotspot: The Balearic Islands as a case study for the Mediterranean region. Biodivers. Conserv. 2023, 32, 2585–2605. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, H.; Hoerandl, E.; de Oliveira Franca, R.; Wang, L.; Hao, J. Autonomous apomixis in Praxelis clematidea (Asteraceae: Eupatorieae), an invasive alien plant. AoB Plants 2021, 13, plab007. [Google Scholar] [CrossRef] [PubMed]
- Maan, I.; Kaur, A.; Sharma, A.; Singh, H.P.; Batish, D.R.; Kohli, R.K.; Arora, N.K. Variations in leaf litter decomposition explain invasion success of Broussonetia papyrifera over confamilial non-invasive Morus alba in urban habitats. Urban For. Urban Green. 2022, 67, 127408. [Google Scholar] [CrossRef]
- Yu, Y.L.; Cheng, H.Y.; Wang, C.Y.; Du, D.L. Heavy drought reduces the decomposition rate of the mixed litters of two composite invasive alien plants. J. Plant Ecol. 2023, 16, rtac047. [Google Scholar] [CrossRef]
- Kumar, M.; Kumar, S.; Verma, A.K.; Joshi, R.K.; Garkoti, S.C. Invasion of Lantana camara and Ageratina adenophora alters the soil physico-chemical characteristics and microbial biomass of chir pine forests in the central Himalaya, India. Catena 2021, 207, 105624. [Google Scholar] [CrossRef]
- Pergl, J.; Vitkova, M.; Hejda, M.; Kutlvasr, J.; Petrik, P.; Sadlo, J.; Vojik, M.; Dvorackova, S.; Fleischhans, R.; Lucanova, A.; et al. Plant-soil interactions in the communities dominated by alien and native plants. Perspect. Plant Ecol. Evol. Syst. 2023, 59, 125721. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G. Effects of exotic plant invasions on soil nutrient cycling processes. Ecosystems 2003, 6, 503–523. [Google Scholar] [CrossRef]
- Prescott, C.E.; Zukswert, J.M. Invasive plant species and litter decomposition: Time to challenge assumptions. New Phytol. 2016, 209, 5–7. [Google Scholar] [CrossRef]
- Dekanova, V.; Svitkova, I.; Novikmec, M.; Svitok, M. Litter breakdown of invasive alien plant species in a pond environment: Rapid decomposition of Solidago canadensis may alter resource dynamics. Limnologica 2021, 90, 125911. [Google Scholar] [CrossRef]
- Hu, X.; Arif, M.; Ding, D.D.; Li, J.J.; He, X.R.; Li, C.X. Invasive plants and species richness impact litter decomposition in Riparian Zones. Front. Plant Sci. 2022, 13, 955656. [Google Scholar] [CrossRef]
- Fu, Y.D.; Xu, W.; Wen, Z.; Han, M.J.; Sun, J.H.; Tang, A.H.; Liu, X. Enhanced atmospheric nitrogen deposition at a rural site in northwest China from 2011 to 2018. Atmos. Res. 2020, 245, 105071. [Google Scholar] [CrossRef]
- Luo, X.S.; Liu, X.J.; Pan, Y.P.; Wen, Z.; Xu, W.; Zhang, L.; Kou, C.L.; Lv, J.L.; Goulding, K. Atmospheric reactive nitrogen concentration and deposition trends from 2011 to 2018 at an urban site in north China. Atmos. Environ. 2020, 224, 117298. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, L.; Liu, X.J.; Li, W.Q.; Lu, S.H.; Zheng, L.X.; Bai, Z.C.; Cai, G.Y.; Zhang, F.S. Atmospheric organic nitrogen deposition in China. Atmos. Environ. 2012, 49, 195–204. [Google Scholar] [CrossRef]
- Zhu, J.X.; Chen, Z.; Wang, Q.F.; Xu, L.; He, N.P.; Jia, Y.L.; Zhang, Q.Y.; Yu, G.R. Potential transition in the effects of atmospheric nitrogen deposition in China. Environ. Pollut. 2020, 258, 113739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Suseela, V. Nitrogen availability modulates the impacts of plant invasion on the chemical composition of soil organic matter. Soil Biol. Biochem. 2021, 156, 108195. [Google Scholar] [CrossRef]
- Zhong, S.S.; Xu, Z.L.; Yu, Y.L.; Cheng, H.Y.; Wang, S.; Wei, M.; Du, D.L.; Wang, C.Y. Acid deposition at higher acidity weakens the antagonistic responses during the co-decomposition of two Asteraceae invasive plants. Ecotoxicol. Environ. Saf. 2022, 243, 114012. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zou, J.; Siemann, E. Interactive effects of elevated CO2 and nitrogen deposition accelerate litter decomposition cycles of invasive tree (Triadica sebifera). For. Ecol. Manag. 2017, 385, 189–197. [Google Scholar] [CrossRef]
- Hernández, E.; Questad, E.J.; Meyer, W.M.; Suding, K.N. The effects of nitrogen deposition and invasion on litter fuel quality and decomposition in a Stipa pulchra grassland. J. Arid Environ. 2019, 162, 35–44. [Google Scholar] [CrossRef]
- Wang, C.Y.; Cheng, H.Y.; Wang, S.; Wei, M.; Du, D.L. Plant community and the influence of plant taxonomic diversity on community stability and invasibility: A case study based on Solidago canadensis L. Sci. Total Environ. 2021, 768, 144518. [Google Scholar] [CrossRef]
- Li, J.Y.; Chen, Y.T.; Guo, Y.Q.; He, Y.X.; Fu, J.W.; Shi, M.Z. Potential suitable areas of Symphyotrichum subulatum based on MaxEnt under future climate scenarios. Plant Prot. 2023, 49, 92–102. [Google Scholar]
- Zhenjiang Bureau of Statistics. Zhenjiang Statistical Yearbook 2022; China Statistics Press: Beijing, China, 2022. [Google Scholar]
- Cornell, S.E. Atmospheric nitrogen deposition: Revisiting the question of the importance of the organic component. Environ. Pollut. 2011, 159, 2214–2222. [Google Scholar] [CrossRef]
- Cornell, S.E.; Jickells, T.D.; Cape, J.N.; Rowland, A.P.; Duce, R.A. Organic nitrogen deposition on land and coastal environments: A review of methods and data. Atmos. Environ. 2003, 37, 2173–2191. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.S. Energy storage and the balance of producers and decomposers in ecological systems. Ecology 1963, 44, 322–331. [Google Scholar] [CrossRef]
- Brookes, P.C.; Landman, A.; Pruden, G.; Jenkinson, D.S. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- GB/T 42485-2023; Soil Quality—Determination of Nitrate, Nitrite and Ammonium in Soils—DExtraction with Potassium Chloride Solution and Determination with Manual Method. National Standard of China 2023; State Administration for Market Regulation of China and Standardization Administration of China: Beijing, China, 2023.
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Wear, E.K.; Wilbanks, E.G.; Nelson, C.E.; Carlson, C.A. Primer selection impacts specific population abundances but not community dynamics in a monthly time-series 16S rRNA gene amplicon analysis of coastal marine bacterioplankton. Environ. Microbiol. 2018, 20, 2709–2726. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Wu, B.D.; Wei, M.; Wang, S.; Rong, X.S.; Du, D.L.; Wang, C.Y. Changes in community structure and metabolic function of soil bacteria depending on the type restoration processing in the degraded alpine grassland ecosystems in Northern Tibet. Sci. Total Environ. 2021, 755, 142619. [Google Scholar] [CrossRef]
- Craine, J.M.; Morrow, C.; Fierer, N. Microbial nitrogen limitation increases decomposition. Ecology 2007, 88, 2105–2113. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Zhou, J.W.; Liu, J.; Jiang, K.; Du, D.L. Responses of soil N-fixing bacteria communities to Amaranthus retroflexus invasion under different forms of N deposition. Agric. Ecosyst. Environ. 2017, 247, 329–336. [Google Scholar] [CrossRef]
- LeBauer, D.S.; Treseder, K.K. Nitrogen limitation of net primary productivity in terrestrial ecosystems is globally distributed. Ecology 2008, 89, 371–379. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Carreiro, M.M.; Repert, D.A. Allocation of extracellular enzymatic activity in relation to litter composition, N deposition, and mass loss. Biogeochemistry 2002, 60, 1–24. [Google Scholar] [CrossRef]
- Lv, Y.; Wang, C.; Wang, F.; Zhao, G.; Pu, G.; Ma, X.; Tian, X. Effects of nitrogen addition on litter decomposition, soil microbial biomass, and enzyme activities between leguminous and non-leguminous forests. Ecol. Res. 2013, 28, 793–800. [Google Scholar] [CrossRef]
- Thirukkumaran, C.M.; Parkinson, D. Microbial respiration, biomass, metabolic quotient and litter decomposition in a lodgepole pine forest floor amended with nitrogen and phosphorous fertilizers. Soil Biol. Biochem. 2000, 32, 59–66. [Google Scholar] [CrossRef]
- Palacin-Lizarbe, C.; Camarero, L.; Hallin, S.; Jones, C.M.; Catalan, J. Denitrification rates in lake sediments of mountains affected by high atmospheric nitrogen deposition. Sci. Rep. 2020, 10, 3003. [Google Scholar] [CrossRef]
- van den Elzen, E.; van den Berg, L.J.L.; van der Weijden, B.; Fritz, C.; Sheppard, L.J.; Lamers, L.P.M. Effects of airborne ammonium and nitrate pollution strongly differ in peat bogs, but symbiotic nitrogen fixation remains unaffected. Sci. Total Environ. 2018, 610–611, 732–740. [Google Scholar] [CrossRef]
- McCrackin, M.L.; Elser, J.J. Atmospheric nitrogen deposition influences denitrification and nitrous oxide production in lakes. Ecology 2010, 91, 528–539. [Google Scholar] [CrossRef]
- Carreiro, M.M.; Sinsabaugh, R.L.; Repert, D.A.; Parkhurst, D.F. Microbial enzyme shifts explain litter decay responses to simulated nitrogen deposition. Ecology 2000, 81, 2359–2365. [Google Scholar] [CrossRef]
- Waldrop, M.P.; Zak, D.R.; Sinsabaugh, R.L.; Gallo, M.; Lauber, C. Nitrogen deposition modifies soil carbon storage through changes in microbial enzymatic activity. Ecol. Appl. 2004, 14, 1172–1177. [Google Scholar] [CrossRef]
- Janssens, I.A.; Dieleman, W.; Luyssaert, S.; Subke, J.A.; Reichstein, M.; Ceulemans, R.; Ciais, P.; Dolman, A.J.; Grace, J.; Matteucci, G.; et al. Reduction of forest soil respiration in response to nitrogen deposition. Nat. Geosci. 2010, 3, 315–322. [Google Scholar] [CrossRef]
- Cuassolo, F.; Diaz Villanueva, V.; Modenutti, B. Litter decomposition of the invasive Potentilla anserina in an invaded and non-invaded freshwater environment of North Patagonia. Biol. Invasions 2020, 22, 1055–1065. [Google Scholar] [CrossRef]
- Liao, C.Z.; Peng, R.H.; Luo, Y.Q.; Zhou, X.H.; Wu, X.W.; Fang, C.M.; Chen, J.K.; Li, B. Altered ecosystem carbon and nitrogen cycles by plant invasion: A meta-analysis. New Phytol. 2008, 177, 706–714. [Google Scholar] [CrossRef]
- Xu, Z.L.; Zhong, S.S.; Yu, Y.L.; Li, Y.; Li, C.; Xu, Z.Y.; Liu, J.; Wang, C.Y.; Du, D.L. Heavy metal contamination alters the co-decomposition of leaves of the invasive tree Rhus typhina L. and the native tree Koelreuteria paniculata Laxm. Plants 2023, 12, 2523. [Google Scholar] [CrossRef] [PubMed]
- Norris, M.D.; Blair, J.M.; Johnson, L.C. Land cover change in eastern Kansas: Litter dynamics of closed-canopy eastern redcedar forests in tallgrass prairie. Can. J. Bot.-Rev. Can. Bot. 2001, 79, 214–222. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G. Ecosystem consequences of biological invasions. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 59–80. [Google Scholar] [CrossRef]
- Vila, M.; Espinar, J.L.; Hejda, M.; Hulme, P.E.; Jarosik, V.; Maron, J.L.; Pergl, J.; Schaffner, U.; Sun, Y.; Pysek, P. Ecological impacts of invasive alien plants: A meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 2011, 14, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Torres, N.; Herrera, I.; Fajardo, L.; Bustamante, R.O. Meta-analysis of the impact of plant invasions on soil microbial communities. BMC Ecol. Evol. 2021, 21, 172. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Staver, A.C. Enhanced activity of soil nutrient-releasing enzymes after plant invasion: A meta-analysis. Ecology 2019, 100, e02830. [Google Scholar] [CrossRef] [PubMed]
- Keet, J.H.; Ellis, A.G.; Hui, C.; Novoa, A.; Le Roux, J.J. Impacts of invasive Australian acacias on soil bacterial community composition, microbial enzymatic activities, and nutrient availability in fynbos soils. Microb. Ecol. 2021, 82, 704–721. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yan, W.B.; Quan, G.M.; Zhang, J.E.; Liang, K.M. Soil microbial carbon utilization, enzyme activities and nutrient availability responses to Bidens pilosa and a non-invasive congener under different irradiances. Sci. Rep. 2017, 7, 11309. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Wang, S.; Wei, M.; Yu, Y.L.; Wang, C.Y. Alien invasive plant Amaranthus spinosus mainly altered the community structure instead of the α diversity of soil N-fixing bacteria under drought. Acta Oecol. 2021, 113, 103788. [Google Scholar] [CrossRef]
- Wang, C.Y.; Wei, M.; Wang, S.; Wu, B.D.; Du, D.L. Cadmium influences the litter decomposition of Solidago canadensis L. and soil N-fixing bacterial communities. Chemosphere 2020, 246, 125717. [Google Scholar] [CrossRef]
- Firmino, V.C.; Brasil, L.S.; Martins, R.T.; Ligeiro, R.; Tonin, A.; Goncalves Junior, J.F.; Juen, L. Litter decomposition of exotic and native plant species of agricultural importance in Amazonian streams. Limnology 2021, 22, 289–297. [Google Scholar] [CrossRef]
- De Castro, W.A.C.; Bonugli-Santos, R.C.; Sibim, A.C.; Da Cunha-Santino, M.B.; Bianchini, I. Enzymatic efficiency of the decomposing microbiota: What does really matter for aquatic macrophytes invasions? Acta Bot. Bras. 2021, 35, 104–110. [Google Scholar] [CrossRef]
| CK | Nitrate | Ammonium | Urea | MixN | |
|---|---|---|---|---|---|
| Decomposition coefficient | 1.54 ± 0.36 b | 2.12 ± 0.16 ab | 2.7 ± 0.5 ab | 3.01 ± 0.59 ab | 3.69 ± 0.45 a |
| Soil pH | 6.8 ± 0.00 ns | 6.83 ± 0.02 ns | 6.77 ± 0.02 ns | 6.76 ± 0.04 ns | 6.74 ± 0.06 ns |
| Soil moisture | 0.38 ± 0.02 ns | 0.41 ± 0.01 ns | 0.42 ± 0.05 ns | 0.43 ± 0.02 ns | 0.44 ± 0.04 ns |
| Soil electrical conductivity | 0.11 ± 0.02 ns | 0.09 ± 0.01 ns | 0.1 ± 0.02 ns | 0.09 ± 0.01 ns | 0.09 ± 0.01 ns |
| Soil organic carbon content | 14.73 ± 1.09 a | 12.86 ± 0.21 b | 13.68 ± 0.47 ab | 13.44 ± 0.42 ab | 13.42 ± 0.38 ab |
| Soil microbial carbon content | 67.81 ± 2.07 ns | 69.16 ± 1.20 ns | 69.3 ± 1.37 ns | 64.5 ± 1.13 ns | 65.63 ± 2.65 ns |
| Soil organic nitrogen content | 4.8 ± 0.32 ns | 4.49 ± 0.27 ns | 4.34 ± 0.24 ns | 4.71 ± 0.18 ns | 4.05 ± 0.29 ns |
| Soil microbial nitrogen content | 8.27 ± 2.78 ns | 8.29 ± 0.49 ns | 9.38 ± 0.45 ns | 8.58 ± 0.62 ns | 9.38 ± 0.65 ns |
| Soil nitrate content | 11.72 ± 0.33 b | 18.43 ± 1.55 a | 14.63 ± 1.34 ab | 18.8 ± 1.64 a | 16.08 ± 1.26 ab |
| Soil ammonium content | 0.87 ± 0.09 a | 0.68 ± 0.04 b | 0.68 ± 0.05 b | 0.65 ± 0.02 b | 0.74 ± 0.06 ab |
| Soil polyphenol oxidase activity | 28.42 ± 4.01 ns | 26.38 ± 2.08 ns | 28.99 ± 2.00 ns | 27.1 ± 1.73 ns | 31.72 ± 2.09 ns |
| Soil FDA hydrolase activity | 3588 ± 664.93 ns | 3027.33 ± 27.00 ns | 2738 ± 202.72 ns | 3020.67 ± 207.84 ns | 2822.67 ± 285.71 ns |
| Soil cellulase activity | 34.67 ± 0.83 ns | 38.78 ± 2.57 ns | 36.3 ± 2.64 ns | 36.45 ± 1.59 ns | 38.31 ± 2.45 ns |
| Soil β-glucosidase activity | 12.24 ± 0.50 ns | 13.48 ± 0.97 ns | 13.23 ± 0.53 ns | 13.37 ± 0.91 ns | 12.57 ± 0.46 ns |
| Soil β-xylosidase activity | 5.67 ± 0.30 a | 5.02 ± 0.13 b | 5.06 ± 0.13 b | 5.11 ± 0.11 b | 5.15 ± 0.12 b |
| Soil sucrase activity | 16.03 ± 4.13 b | 25.41 ± 3.90 ab | 16.99 ± 2.73 ab | 29.82 ± 5.56 a | 21.41 ± 2.87 ab |
| Soil protease activity | 0.38 ± 0.01 ns | 0.37 ± 0.02 ns | 0.36 ± 0.02 ns | 0.37 ± 0.03 ns | 0.37 ± 0.01 ns |
| Soil urease activity | 90.13 ± 2.23 ns | 99.95 ± 5.86 ns | 99.3 ± 4.63 ns | 112.44 ± 7.23 ns | 92.16 ± 11.48 ns |
| OTU’s species index | 3036.67 ± 323.39 b | 3504.78 ± 136.46 a | 3741.55 ± 104.60 a | 3582.17 ± 131.50 a | 3674 ± 72.82 a |
| Shannon’s diversity index | 8.95 ± 0.17 ns | 9.02 ± 0.15 ns | 9.14 ± 0.11 ns | 9.25 ± 0.04 ns | 9.23 ± 0.10 ns |
| Simpson’s dominance index | 0.99 ± 0.00 ns | 0.99 ± 0.00 ns | 0.99 ± 0.00 ns | 0.99 ± 0.00 ns | 0.99 ± 0.00 ns |
| Pielou’s evenness index | 0.78 ± 0.01 ns | 0.77 ± 0.01 ns | 0.77 ± 0.01 ns | 0.79 ± 0.00 ns | 0.78 ± 0.01 ns |
| Chao1’s richness index | 3563.04 ± 246.47 b | 4132.47 ± 138.46 a | 4338.73 ± 121.98 a | 4153.33 ± 170.35 a | 4289.46 ± 75.54 a |
| ACE’s richness index | 3692.02 ± 246.08 b | 4363.74 ± 147.94 a | 4587.5 ± 131.23 a | 4352.47 ± 191.36 a | 4511.73 ± 84.26 a |
| Phylogenetic diversity index | 368.45 ± 33.46 b | 421.12 ± 13.84 a | 446.85 ± 10.13 a | 432.94 ± 9.49 a | 435.39 ± 6.63 a |
| PL | BP | CC | SC | SS | |
|---|---|---|---|---|---|
| Decomposition coefficient | 2.94 ± 0.67 ab | 2.74 ± 0.48 ab | 3.37 ± 0.57 a | 2.37 ± 0.39 ab | 1.65 ± 0.23 b |
| Soil pH | 6.82 ± 0.03 ns | 6.75 ± 0.03 ns | 6.79 ± 0.03 ns | 6.84 ± 0.01 ns | 6.78 ± 0.04 ns |
| Soil moisture | 0.47 ± 0.04 a | 0.43 ± 0.02 ab | 0.41 ± 0.03 ab | 0.35 ± 0.02 b | 0.36 ± 0.03 b |
| Soil electrical conductivity | 0.08 ± 0.01 ab | 0.1 ± 0.01 a | 0.09 ± 0.00 a | 0.05 ± 0.01 b | 0.08 ± 0.01 ab |
| Soil organic carbon content | 13.38 ± 0.46 ns | 12.86 ± 0.20 ns | 13.5 ± 0.50 ns | 13.02 ± 0.53 ns | 13.28 ± 0.40 ns |
| Soil microbial carbon content | 68.95 ± 1.93 ns | 66.51 ± 1.07 ns | 67.7 ± 2.09 ns | 65.3 ± 1.34 ns | 64.9 ± 2.64 ns |
| Soil organic nitrogen content | 4.33 ± 0.29 ns | 4.4 ± 0.26 ns | 4.33 ± 0.34 ns | 4.81 ± 0.18 ns | 4.32 ± 0.20 ns |
| Soil microbial nitrogen content | 8.56 ± 0.96 ns | 9.49 ± 0.68 ns | 8.82 ± 0.74 ns | 7.89 ± 0.57 ns | 8.76 ± 0.55 ns |
| Soil nitrate content | 18.96 ± 2.44 ns | 15.8 ± 2.87 ns | 15.52 ± 2.46 ns | 14.26 ± 2.11 ns | 12.12 ± 1.59 ns |
| Soil ammonium content | 0.7 ± 0.07 ns | 0.66 ± 0.05 ns | 0.75 ± 0.06 ns | 0.62 ± 0.03 ns | 0.66 ± 0.02 ns |
| Soil polyphenol oxidase activity | 24.96 ± 1.71 b | 29.57 ± 2.56 ab | 33.47 ± 0.77 a | 29 ± 1.99 ab | 31.36 ± 1.30 a |
| Soil FDA hydrolase activity | 2839.2 ± 178.87 a | 3168 ± 154.72 a | 3171.2 ± 258.25 a | 2888.8 ± 273.47 a | 2074 ± 154.99 b |
| Soil cellulase activity | 35.55 ± 1.24 ns | 39.71 ± 2.31 ns | 42.74 ± 3.75 ns | 38.69 ± 4.06 ns | 34.68 ± 0.56 ns |
| Soil β-glucosidase activity | 13.21 ± 0.50 ab | 13.73 ± 0.82 a | 13.99 ± 0.66 a | 13.01 ± 1.15 ab | 11.08 ± 0.41 b |
| Soil β-xylosidase activity | 5.24 ± 0.13 ns | 4.99 ± 0.09 ns | 5.2 ± 0.06 ns | 4.99 ± 0.18 ns | 5.11 ± 0.16 ns |
| Soil sucrase activity | 25.54 ± 2.23 ab | 33.7 ± 2.75 a | 21.44 ± 2.85 bc | 14.83 ± 4.17 c | 13.66 ± 1.80 c |
| Soil protease activity | 0.36 ± 0.01 ns | 0.4 ± 0.03 ns | 0.41 ± 0.02 ns | 0.37 ± 0.03 ns | 0.37 ± 0.02 ns |
| Soil urease activity | 90.23 ± 12.43 ns | 97.82 ± 6.17 ns | 111.45 ± 4.42 ns | 110.67 ± 9.74 ns | 97.92 ± 8.08 ns |
| OTU’s species index | 3603.93 ± 114.99 ns | 3847.13 ± 97.5 ns | 3740.8 ± 53.99 ns | 3594.33 ± 124.32 ns | 3658.5 ± 129.07 ns |
| Shannon’s diversity index | 9.08 ± 0.14 ns | 9.24 ± 0.13 ns | 9.25 ± 0.06 ns | 9.12 ± 0.08 ns | 9.27 ± 0.05 ns |
| Simpson’s dominance index | 0.99 ± 0.00 ns | 0.99 ± 0.00 ns | 0.99 ± 0.00 ns | 0.99 ± 0.00 ns | 0.99 ± 0.00 ns |
| Pielou’s evenness index | 0.77 ± 0.01 ns | 0.78 ± 0.01 ns | 0.78 ± 0.01 ns | 0.77 ± 0.01 ns | 0.79 ± 0.00 ns |
| Chao1’s richness index | 4144.24 ± 128.64 ns | 4477.88 ± 94.19 ns | 4407.77 ± 66.63 ns | 4232.75 ± 117.91 ns | 4292.45 ± 162.42 ns |
| ACE’s richness index | 4342.18 ± 141.83 ns | 4703.6 ± 104.27 ns | 4671.17 ± 83.87 ns | 4475.48 ± 127.43 ns | 4546.44 ± 185.6 ns |
| Phylogenetic diversity index | 426.74 ± 11.13 ab | 457.37 ± 9.50 a | 444.33 ± 4.62 ab | 422.4 ± 11.78 b | 438.97 ± 10.28 ab |
| k | k | k | ||||||
|---|---|---|---|---|---|---|---|---|
| Soil pH | r | 0.045 | Soil ammonium content | r | 0.004 | Soil urease activity | r | 0.291 * |
| p | 0.699 | p | 0.972 | p | 0.011 | |||
| Soil moisture | r | 0.175 | Soil polyphenol oxidase activity | r | 0.102 | OTU’s species index | r | 0.012 |
| p | 0.133 | p | 0.385 | p | 0.919 | |||
| Soil electrical conductivity | r | 0.239 * | Soil FDA hydrolase activity | r | 0.386 *** | Shannon’s diversity index | r | 0.179 |
| p | 0.039 | p | <0.001 | p | 0.125 | |||
| Soil organic carbon content | r | 0.088 | Soil cellulase activity | r | 0.034 | Simpson’s dominance index | r | 0.167 |
| p | 0.452 | p | 0.771 | p | 0.153 | |||
| Soil microbial carbon content | r | −0.128 | Soil β-glucosidase activity | r | 0.256 * | Pielou’s evenness index | r | 0.195 |
| p | 0.273 | p | 0.026 | p | 0.094 | |||
| Soil organic nitrogen content | r | −0.016 | Soil β-xylosidase activity | r | 0.296 ** | Chao1’s richness index | r | 0.015 |
| p | 0.892 | p | 0.010 | p | 0.899 | |||
| Soil microbial nitrogen content | r | 0.005 | Soil sucrase activity | r | 0.108 | ACE’s richness index | r | 0.002 |
| p | 0.969 | p | 0.359 | p | 0.986 | |||
| Soil nitrate content | r | 0.375 *** | Soil protease activity | r | 0.093 | Phylogenetic diversity index | r | −0.025 |
| p | <0.001 | p | 0.427 | p | 0.831 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Li, Y.; Zhong, S.; Xu, Z.; Xu, Z.; Zhu, M.; Wei, Y.; Wang, C.; Du, D. Do the Leaves of Multiple Invasive Plants Decompose More Easily than a Native Plant’s under Nitrogen Deposition with Different Forms? Nitrogen 2024, 5, 202-218. https://doi.org/10.3390/nitrogen5010014
Li C, Li Y, Zhong S, Xu Z, Xu Z, Zhu M, Wei Y, Wang C, Du D. Do the Leaves of Multiple Invasive Plants Decompose More Easily than a Native Plant’s under Nitrogen Deposition with Different Forms? Nitrogen. 2024; 5(1):202-218. https://doi.org/10.3390/nitrogen5010014
Chicago/Turabian StyleLi, Chuang, Yue Li, Shanshan Zhong, Zhelun Xu, Zhongyi Xu, Mawei Zhu, Yuqing Wei, Congyan Wang, and Daolin Du. 2024. "Do the Leaves of Multiple Invasive Plants Decompose More Easily than a Native Plant’s under Nitrogen Deposition with Different Forms?" Nitrogen 5, no. 1: 202-218. https://doi.org/10.3390/nitrogen5010014
APA StyleLi, C., Li, Y., Zhong, S., Xu, Z., Xu, Z., Zhu, M., Wei, Y., Wang, C., & Du, D. (2024). Do the Leaves of Multiple Invasive Plants Decompose More Easily than a Native Plant’s under Nitrogen Deposition with Different Forms? Nitrogen, 5(1), 202-218. https://doi.org/10.3390/nitrogen5010014








