Nitrogen-Fixing Symbiotic Paraburkholderia Species: Current Knowledge and Future Perspectives
Abstract
1. Introduction
2. Two Decades since the Discovery of Nitrogen-Fixing Symbiotic Beta-proteobacteria
| P. phymatum Strains | Sources (Country) | Plant Host | Publication |
|---|---|---|---|
| STM815T | French Guiana | Machaerium lunatum/ Mimosa pudica * | Moulin et al., 2001 [10]/ Mishra et al., 2012 [48] |
| GR01, GR03, GR05, GR06 1,2 | Morocco | Phaseolus vulgaris | Talbi et al., 2010 [49] |
| NGR114 1,3 | Papua New Guinea | M. pudica | Elliott et al., 2007a [35] |
| NGR195A 1,3 | Papua New Guinea | M. invisa | Elliott et al., 2007a [35] |
| STM3619, STM3622, STM3623, STM3631, STM3665, STM3666, STM3667, STM3668, STM3669, STM3674, STM3675, STM3676, STM4205, STM4207, STM4208, STM4211, STM4212, STM4214, STM4216, STM4217, STM4219, STM4221, STM4223, STM4225, STM4302, STM4303, STM4305, STM4308, STM4312, STM4313, STM4316, STM4317, STM4318, STM4319, STM4320, STM4322, STM4323, STM4324, STM4325, STM4326, STM4327, STM4328, STM4332, STM4333, STM4334, STM4335, STM4337, STM4338, STM4339, STM4342, STM4343, STM4344, STM4345, STM4346, STM4337, STM6016, STM6019, STM6023, STM6028, STM6025, STM6027, STM6031, STM6017, STM6022 1,4 | French Guiana | M. pudica | Mishra et al., 2012 [48] |
| SWF66029, SWF66286 1,4 | China | M. pudica | Liu et al., 2011 [50] |
| SWF67297 1,4 | China | M. pudica | Liu et al., 2012 [51] |
| MP20, MPJ1 1 | India | M. pudica | Gehlot et al., 2013 [52] |
| CVRDII_2 1,5 | Brazil | Parapiptadenia pterosperma | Bournaud et al., 2013 [53] |
| STM3714 1,4 | Guinea | M. pudica | Melkonian et al., 2014 [7] |
| HBU52006, HBU52001, HBU35004, and 52 other strains 1,4 | China | M. pudica | Liu et al., 2020 [54] |
| HBU67642, HBU67643 1,4 | China | M. diplotricha | Liu et al., 2020 [54] |
3. Geographical Distribution and Phylogeny of Paraburkholderia Species
4. High Competitiveness of Paraburkholderia in the Rhizosphere
4.1. Abiotic Factors and Stress Responses
4.2. Competition between Soil Bacteria for Survival and Root Nodulation
4.2.1. Exploitative or Passive Competition
4.2.2. Interference or Active Competition
5. Interaction with the Plant Host
5.1. “Opening the Gate”: Rhizobium–Legume Recognition
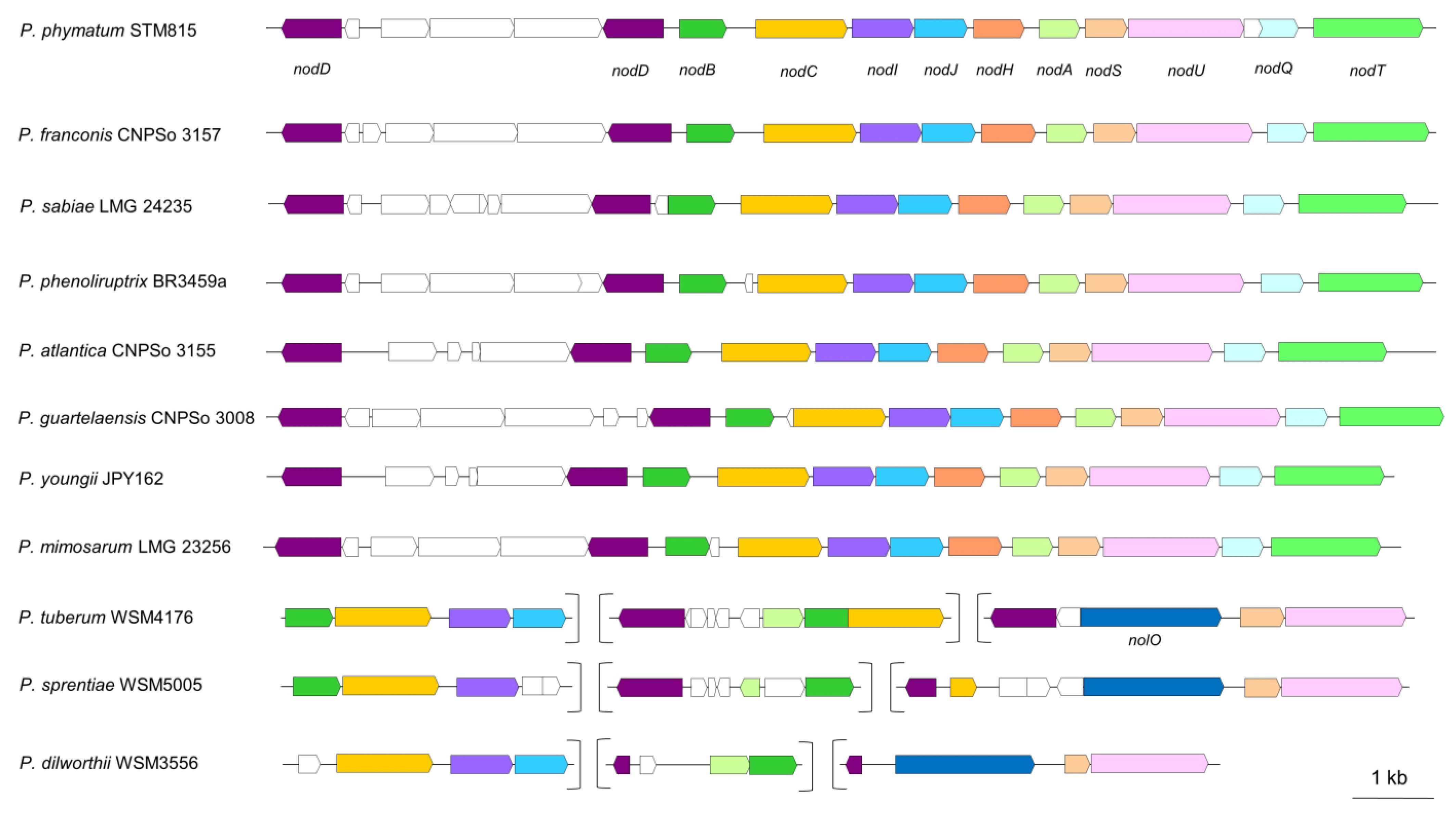
5.2. The Steps towards Nodule Development
5.3. “Achieving the Goal”: Differentiation to BNF-Performing Bacteroids
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galloway, J.N.; Aber, J.D.; Erisman, J.W.; Seitzinger, S.P.; Howarth, R.W.; Cowling, E.B.; Cosby, B.J. The Nitrogen Cascade. Bioscience 2003, 53, 341–356. [Google Scholar] [CrossRef]
- Bloom, A.J. The Increasing Importance of Distinguishing among Plant Nitrogen Sources. Curr. Opin. Plant Biol. 2015, 25, 10–16. [Google Scholar] [CrossRef]
- Gruber, N.; Galloway, J.N. An Earth-System Perspective of the Global Nitrogen Cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef]
- Bottomley, P.J.; Myrold, D.D. Biological N Inputs. In Soil Microbiology, Ecology and Biochemistry; Elsevier: Oxford, UK, 2007; pp. 365–387. [Google Scholar]
- Dixon, R.; Kahn, D. Genetic Regulation of Biological Nitrogen Fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef]
- Willems, A. The Taxonomy of Rhizobia: An Overview. Plant Soil 2006, 287, 3–14. [Google Scholar] [CrossRef]
- Melkonian, R.; Moulin, L.; Béna, G.; Tisseyre, P.; Chaintreuil, C.; Heulin, K.; Rezkallah, N.; Klonowska, A.; Gonzalez, S.; Simon, M.; et al. The Geographical Patterns of Symbiont Diversity in the Invasive Legume Mimosa pudica Can Be Explained by the Competitiveness of Its Symbionts and by the Host Genotype. Environ. Microbiol. 2014, 16, 2099–2111. [Google Scholar] [CrossRef]
- Sachs, J.L.; Quides, K.W.; Wendlandt, C.E. Legumes versus Rhizobia: A Model for Ongoing Conflict in Symbiosis. New Phytol. 2018, 219, 1199–1206. [Google Scholar] [CrossRef]
- Lindström, K.; Mousavi, S.A. Effectiveness of Nitrogen Fixation in Rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef]
- Moulin, L.; Munive, A.; Dreyfus, B.; Boivin-Masson, C. Nodulation of Legumes by Members of the Beta-Subclass of Proteobacteria. Nature 2001, 411, 948–950. [Google Scholar] [CrossRef]
- Young, J.P.W.; Johnston, A.W.B. The Evolution of Specificity in the Legume-Rhizobium Symbiosis. Tree 1989, 4, 341–349. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Dwivedi, S.L.; Ambrose, M.; Ellis, N.; Berger, J.; Smýkal, P.; Debouck, D.; Duc, G.; Dumet, D.; Flavell, A. Legume Genetic Resources: Management, Diversity Assessment, and Utilization in Crop Improvement. Euphytica 2011, 180, 27–47. [Google Scholar] [CrossRef]
- Sprent, J.I.; Ardley, J.; James, E.K. Biogeography of Nodulated Legumes and Their Nitrogen-Fixing Symbionts. New Phytol. 2017, 215, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Griesmann, M.; Chang, Y.; Liu, X.; Song, Y.; Haberer, G.; Crook, M.B.; Billault-Penneteau, B.; Lauressergues, D.; Keller, J.; Imanishi, L.; et al. Phylogenomics Reveals Multiple Losses of Nitrogen-Fixing Root Nodule Symbiosis. Science 2018, 361, aat1743. [Google Scholar] [CrossRef] [PubMed]
- van Velzen, R.; Doyle, J.J.; Geurts, R. A Resurrected Scenario: Single Gain and Massive Loss of Nitrogen-Fixing Nodulation. Trends Plant Sci. 2019, 24, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Sprent, J.I. Evolving Ideas of Legume Evolution and Diversity: A Taxonomic Perspective on the Occurrence of Nodulation. New Phytol. 2007, 174, 11–25. [Google Scholar] [CrossRef]
- Walker, R.; Agapakis, C.M.; Watkin, E.; Hirsch, A.M. Symbiotic Nitrogen Fixation in Legumes: Perspectives on the Diversity and Evolution of Nodulation by Rhizobium and Burkholderia Species. In Biological Nitrogen Fixation; Wiley Blackwell: Hoboken, NJ, USA, 2015; Volume 2, pp. 913–925. ISBN 9781119053095. [Google Scholar]
- Gyaneshwar, P.; Hirsch, A.M.; Moulin, L.; Chen, W.M.; Elliott, G.N.; Bontemps, C.; de Los Santos, P.E.; Gross, E.; dos Reis, F.B.; Janet, I.S.; et al. Legume-Nodulating Betaproteobacteria: Diversity, Host Range, and Future Prospects. Mol. Plant-Microbe Interact. 2011, 24, 1276–1288. [Google Scholar] [CrossRef]
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.R.; Pinto, R.B.; Boatwright, J.S.; Borges, L.M.; Brown, G.K.; Bruneau, A.; et al. A New Subfamily Classification of the Leguminosae Based on a Taxonomically Comprehensive Phylogeny. Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef]
- Paulitsch, F.; Delamuta, J.R.M.; Ribeiro, R.A.; da Silva Batista, J.S.; Hungria, M. Phylogeny of Symbiotic Genes Reveals Symbiovars within Legume-Nodulating Paraburkholderia Species. Syst. Appl. Microbiol. 2020, 43, 126151. [Google Scholar] [CrossRef]
- de Faria, S.M.; Ringelberg, J.J.; Gross, E.; Koenen, E.J.M.; Cardoso, D.; Ametsitsi, G.K.D.; Akomatey, J.; Maluk, M.; Tak, N.; Gehlot, H.S.; et al. The Innovation of the Symbiosome Has Enhanced the Evolutionary Stability of Nitrogen Fixation in Legumes. New Phytol. 2022, 235, 2365–2377. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E.; Bedmar, E.J. Bacterial Associations with Legumes. CRC Crit. Rev. Plant Sci. 2015, 34, 17–42. [Google Scholar] [CrossRef]
- FAOSTAT FAO Statistical Databases. Food and Agriculture Organization of the United Nations. 2021. Available online: https://www.fao.org/faostat/en/#data/qcl (accessed on 6 February 2023).
- Velázquez, E.; García-Fraile, P.; Ramírez-Bahena, M.H.; Rivas, R.; Martínez-Molina, E. Current Status of the Taxonomy of Bacteria Able to Establish Nitrogen-Fixing Legume Symbiosis. In Microbes for Legume Improvement, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–43. ISBN 9783319591742. [Google Scholar]
- Vandamme, P.; Goris, J.; Chen, W.M.; de Vos, P.; Willems, A. Burkholderia tuberum sp. Nov. and Burkholderia phymatum sp. Nov., Nodulate the Roots of Tropical Legumes. Syst. Appl. Microbiol. 2002, 25, 507–512. [Google Scholar] [CrossRef]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular Signatures and Phylogenomic Analysis of the Genus Burkholderia: Proposal for Division of This Genus into the Emended Genus Burkholderia Containing Pathogenic Organisms and a New Genus Paraburkholderia Gen. Nov. Harboring Environmental Species. Front. Genet. 2014, 5, 00429. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; Laevens, S.; Lee, T.M.; Coenye, T.; de Vos, P.; Mergeay, M.; Vandamme, P. Ralstonia taiwanensis sp. Nov., Isolated from Root Nodules of Mimosa Species and Sputum of a Cystic Fibrosis Patient. Int. J. Syst. Evol. Microbiol. 2001, 51, 1729–1735. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, P.; Coenye, T. Taxonomy of the Genus Cupriavidus: A Tale of Lost and Found. Int. J. Syst. Evol. Microbiol. 2004, 54, 2285–2289. [Google Scholar] [CrossRef]
- Bontemps, C.; Elliott, G.N.; Simon, M.F.; dos Reis Júnior, F.B.; Gross, E.; Lawton, R.C.; Neto, N.E.; de Fátima Loureiro, M.; de Faria, S.M.; Sprent, J.I.; et al. Burkholderia Species Are Ancient Symbionts of Legumes. Mol. Ecol. 2010, 19, 44–52. [Google Scholar] [CrossRef]
- Chen, W.-M.; James, E.K.; Prescott, A.R.; Kierans, M.; Sprent, J.I. Nodulation of Mimosa spp. by the β-Proteobacterium Ralstonia Taiwanensis. MPMI 2003, 16, 1051–1061. [Google Scholar] [CrossRef]
- Chen, W.; de Faria, S.M.; Pitard, R.M.; Simo, J.L.; Prescott, A.R.; Elliott, G.N.; Sprent, J.I.; Young, J.P.W.; James, E.K. Proof That Burkholderia Strains Form Effective Symbioses with Legumes: A Study of Novel Mimosa-Nodulating Strains from South America. Appl. Environ. Microbiol. 2005, 71, 7461–7471. [Google Scholar] [CrossRef]
- Chen, W.M.; James, E.K.; Chou, J.H.; Sheu, S.Y.; Yang, S.Z.; Sprent, J.I. β-Rhizobia from Mimosa Pigra, a Newly Discovered Invasive Plant in Taiwan. New Phytol. 2005, 168, 661–675. [Google Scholar] [CrossRef]
- Chen, W.M.; de Faria, S.M.; James, E.K.; Elliott, G.N.; Lin, K.Y.; Chou, J.H.; Sheu, S.Y.; Cnockaert, M.; Sprent, J.I.; Vandamme, P. Burkholderia Nodosa sp. Nov., Isolated from Root Nodules of the Woody Brazilian Legumes Mimosa Bimucronata and Mimosa Scabrella. Int. J. Syst. Evol. Microbiol. 2007, 57, 1055–1059. [Google Scholar] [CrossRef]
- Chen, W.M.; James, E.K.; Coenye, T.; Chou, J.H.; Barrios, E.; de Faria, S.M.; Elliott, G.N.; Sheu, S.Y.; Sprent, J.I.; Vandamme, P. Burkholderia Mimosarum sp. Nov., Isolated from Root Nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 2006, 56, 1847–1851. [Google Scholar] [CrossRef] [PubMed]
- Elliott, G.N.; Chen, W.M.; Chou, J.H.; Wang, H.C.; Sheu, S.Y.; Perin, L.; Reis, V.M.; Moulin, L.; Simon, M.F.; Bontemps, C.; et al. Burkholderia phymatum Is a Highly Effective Nitrogen-Fixing Symbiont of Mimosa spp. and Fixes Nitrogen Ex Planta. New Phytol. 2007, 173, 168–180. [Google Scholar] [CrossRef]
- Elliott, G.N.; Chen, W.M.; Bontemps, C.; Chou, J.H.; Young, J.P.W.; Sprent, J.I.; James, E.K. Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann. Bot. 2007, 100, 1403–1411. [Google Scholar] [CrossRef]
- Reis, V.M.; Estrada-De Los Santos, P.; Tenorio-Salgado, S.; Vogel, J.; Stoffels, M.; Guyon, S.; Mavingui, P.; Baldani, V.L.D.; Schmid, M.; Baldani, J.I. Burkholderia tropica sp. Nov., a Novel Nitrogen-Fixing, Plant-Associated Bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 2155–2162. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Mellado, J.; Martínez-Aguilar, L.; Paredes-Valdez, G.; Estrada-De Los Santos, P. Burkholderia unamae sp. Nov., an N2-Fixing Rhizospheric and Endophytic Species. Int. J. Syst. Evol. Microbiol. 2004, 54, 1165–1172. [Google Scholar] [CrossRef]
- Coenye, T.; Vandamme, P. Diversity and Significance of Burkholderia Species Occupying Diverse Ecological Niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Angus, A.A.; Agapakis, C.M.; Fong, S.; Yerrapragada, S.; Estrada-de Los Santos, P.; Yang, P.; Song, N.; Kano, S.; Caballero-Mellado, J.; de Faria, S.M.; et al. Plant-Associated Symbiotic Burkholderia Species Lack Hallmark Strategies Required in Mammalian Pathogenesis. PLoS ONE 2014, 9, e83779. [Google Scholar] [CrossRef]
- Eberl, L.; Vandamme, P. Members of the Genus Burkholderia: Good and Bad Guys. F1000Research 2016, 5, 1007. [Google Scholar] [CrossRef] [PubMed]
- Dobritsa, A.P.; Samadpour, M. Transfer of Eleven Species of the Genus Burkholderia to the Genus Paraburkholderia and Proposal of Caballeronia Gen. Nov. to Accommodate Twelve Species of the Genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016, 66, 2836–2846. [Google Scholar] [CrossRef]
- Beukes, C.W.; Palmer, M.; Manyaka, P.; Chan, W.Y.; Avontuur, J.R.; van Zyl, E.; Huntemann, M.; Clum, A.; Pillay, M.; Palaniappan, K.; et al. Genome Data Provides High Support for Generic Boundaries in Burkholderia Sensu Lato. Front. Microbiol. 2017, 8, 1154. [Google Scholar] [CrossRef]
- Estrada-de los Santos, P.; Palmer, M.; Chávez-Ramírez, B.; Beukes, C.; Steenkamp, E.T.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E.; et al. Whole Genome Analyses Suggests That Burkholderia Sensu Lato Contains Two Additional Novel Genera (Mycetohabitans Gen. Nov., and Trinickia Gen. Nov.): Implications for the Evolution of Diazotrophy and Nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef]
- Lin, Q.; Lv, Y.; Gao, Z.; Qiu, L. Pararobbsia silviterrae Gen. Nov., sp. Nov., Isolated from Forest Soil and Reclassification of Burkholderia alpina as Pararobbsia alpina Comb. Nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 1412–1420. [Google Scholar] [CrossRef]
- Rahimlou, S.; Bahram, M.; Tedersoo, L. Phylogenomics Reveals the Evolution of Root Nodulating Alpha- and Beta-Proteobacteria (Rhizobia). Microbiol. Res. 2021, 250, 126788. [Google Scholar] [CrossRef]
- Moulin, L.; Klonowska, A.; Caroline, B.; Booth, K.; Vriezen, J.A.C.; Melkonian, R.; James, E.K.; Young, J.P.W.; Bena, G.; Hauser, L.; et al. Complete Genome Sequence of Burkholderia phymatum STM815T, a Broad Host Range and Efficient Nitrogen-Fixing Symbiont of Mimosa Species. Stand. Genom. Sci. 2014, 9, 763–774. [Google Scholar] [CrossRef]
- Mishra, R.P.N.; Tisseyre, P.; Melkonian, R.; Chaintreuil, C.; Miché, L.; Klonowska, A.; Gonzalez, S.; Bena, G.; Laguerre, G.; Moulin, L. Genetic Diversity of Mimosa Pudica Rhizobial Symbionts in Soils of French Guiana: Investigating the Origin and Diversity of Burkholderia phymatum and Other Beta-Rhizobia. FEMS Microbiol. Ecol. 2012, 79, 487–503. [Google Scholar] [CrossRef]
- Talbi, C.; Delgado, M.J.; Girard, L.; Ramírez-Trujillo, A.; Caballero-Mellado, J.; Bedmar, E.J. Burkholderia phymatum Strains Capable of Nodulating Phaseolus Vulgaris Are Present in Moroccan Soils. Appl. Environ. Microbiol. 2010, 76, 4587–4591. [Google Scholar] [CrossRef]
- Liu, X.Y.; Wu, W.; Wang, E.T.; Zhang, B.; Macdermott, J.; Chen, W.X. Phylogenetic Relationships and Diversity of β-Rhizobia Associated with Mimosa Species Grown in Sishuangbanna, China. Int. J. Syst. Evol. Microbiol. 2011, 61, 334–342. [Google Scholar] [CrossRef]
- Liu, X.; Wei, S.; Wang, F.; James, E.K.; Guo, X.; Zagar, C.; Xia, L.G.; Dong, X.; Wang, Y.P. Burkholderia and Cupriavidus spp. Are the Preferred Symbionts of Mimosa spp. in Southern China. FEMS Microbiol. Ecol. 2012, 80, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Gehlot, H.S.; Tak, N.; Kaushik, M.; Mitra, S.; Chen, W.M.; Poweleit, N.; Panwar, D.; Poonar, N.; Parihar, R.; Tak, A.; et al. An Invasive Mimosa in India Does Not Adopt the Symbionts of Its Native Relatives. Ann. Bot. 2013, 112, 179–196. [Google Scholar] [CrossRef]
- Bournaud, C.; de Faria, S.M.; dos Santos, J.M.F.; Tisseyre, P.; Silva, M.; Chaintreuil, C.; Gross, E.; James, E.K.; Prin, Y.; Moulin, L. Burkholderia Species Are the Most Common and Preferred Nodulating Symbionts of the Piptadenia Group (Tribe Mimoseae). PLoS ONE 2013, 8, 0063478. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; You, S.; Liu, H.; Yuan, B.; Wang, H.; James, E.K.; Wang, F.; Cao, W.; Liu, Z.K. Diversity and Geographic Distribution of Microsymbionts Associated with Invasive Mimosa Species in Southern China. Front. Microbiol. 2020, 11, 563389. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.F.; Parker, M.A. Prevalence of Burkholderia sp. Nodule Symbionts on Four Mimosoid Legumes from Barro Colorado Island, Panama. Syst. Appl. Microbiol. 2005, 28, 57–65. [Google Scholar] [CrossRef]
- Barrett, C.F.; Parker, M.A. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. Nodule Bacteria on Two Mimosa spp. in Costa Rica. Appl. Environ. Microbiol. 2006, 72, 1198–1206. [Google Scholar] [CrossRef]
- Garau, G.; Yates, R.J.; Deiana, P.; Howieson, J.G. Novel Strains of Nodulating Burkholderia Have a Role in Nitrogen Fixation with Papilionoid Herbaceous Legumes Adapted to Acid, Infertile Soils. Soil Biol. Biochem. 2009, 41, 125–134. [Google Scholar] [CrossRef]
- Beukes, C.W.; Venter, S.N.; Law, I.J.; Phalane, F.L.; Steenkamp, E.T. South African Papilionoid Legumes Are Nodulated by Diverse Burkholderia with Unique Nodulation and Nitrogen-Fixation Loci. PLoS ONE 2013, 8, 0068406. [Google Scholar] [CrossRef]
- Howieson, J.G.; de Meyer, S.E.; Vivas-Marfisi, A.; Ratnayake, S.; Ardley, J.K.; Yates, R.J. Novel Burkholderia Bacteria Isolated from Lebeckia ambigua—A Perennial Suffrutescent Legume of the Fynbos. Soil. Biol. Biochem. 2013, 60, 55–64. [Google Scholar] [CrossRef]
- Liu, W.Y.Y.; Ridgway, H.J.; James, T.K.; James, E.K.; Chen, W.M.; Sprent, J.I.; Young, J.P.W.; Andrews, M. Burkholderia sp. Induces Functional Nodules on the South African Invasive Legume Dipogon lignosus (Phaseoleae) in New Zealand Soils. Microb. Ecol. 2014, 68, 542–555. [Google Scholar] [CrossRef]
- Lemaire, B.; Dlodlo, O.; Chimphango, S.; Stirton, C.; Schrire, B.; Boatwright, J.S.; Honnay, O.; Smets, E.; Sprent, J.; James, E.K.; et al. Symbiotic Diversity, Specificity and Distribution of Rhizobia in Native Legumes of the Core Cape Subregion (South Africa). FEMS Microbiol. Ecol. 2015, 91, fiu024. [Google Scholar] [CrossRef]
- Lemaire, B.; van Cauwenberghe, J.; Verstraete, B.; Chimphango, S.; Stirton, C.; Honnay, O.; Smets, E.; Sprent, J.; James, E.K.; Muasya, A.M. Characterization of the Papilionoid-Burkholderia Interaction in the Fynbos Biome: The Diversity and Distribution of Beta-Rhizobia Nodulating Podalyria calyptrata (Fabaceae, Podalyrieae). Syst. Appl. Microbiol. 2016, 39, 41–48. [Google Scholar] [CrossRef]
- Lemaire, B.; Chimphango, S.B.M.; Stirton, C.; Rafudeen, S.; Honnay, O.; Smets, E.; Chen, W.M.; Sprent, J.; James, E.K.; Muasya, A.M. Biogeographical Patterns of Legume-Nodulating Burkholderia spp.: From African Fynbos to Continental Scales. Appl Environ. Microbiol. 2016, 82, 5099–5115. [Google Scholar] [CrossRef]
- Beukes, C.W.; Boshoff, F.S.; Phalane, F.L.; Hassen, A.I.; le Roux, M.M.; Stȩpkowski, T.; Venter, S.N.; Steenkamp, E.T. Both Alpha- and Beta-Rhizobia Occupy the Root Nodules of Vachellia Karroo in South Africa. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Mavima, L.; Beukes, C.W.; Palmer, M.; de Meyer, S.E.; James, E.K.; Maluk, M.; Muasya, M.A.; Avontuur, J.R.; Chan, W.Y.; Venter, S.N. Delineation of Paraburkholderia tuberum sensu stricto and Description of Paraburkholderia podalyriae sp. Nov. Nodulating the South African Legume Podalyria calyptrata. Syst. Appl. Microbiol. 2022, 45, 126316. [Google Scholar] [CrossRef]
- Parker, M.A.; Wurtz, A.K.; Paynter, Q. Nodule Symbiosis of Invasive Mimosa pigra in Australia and in Ancestral Habitats: A Comparative Analysis. Biol. Invasions 2007, 9, 127–138. [Google Scholar] [CrossRef]
- Sheu, S.Y.; Chen, M.H.; Liu, W.Y.Y.; Andrews, M.; James, E.K.; Ardley, J.K.; de Meyer, S.E.; James, T.K.; Howieson, J.G.; Coutinho, B.G.; et al. Burkholderia dipogonis sp. Nov, Isolated from Root Nodules of Dipogon lignosus in New Zealand and Western Australia. Int. J. Syst. Evol. Microbiol. 2015, 65, 4716–4723. [Google Scholar] [CrossRef]
- Elliott, G.N.; Chou, J.H.; Chen, W.M.; Bloemberg, G.V.; Bontemps, C.; Martínez-Romero, E.; Velázquez, E.; Young, J.P.W.; Sprent, J.I.; James, E.K. Burkholderia spp. Are the Most Competitive Symbionts of Mimosa, Particularly under N-Limited Conditions. Environ. Microbiol. 2009, 11, 762–778. [Google Scholar] [CrossRef]
- dos Reis Junior, F.B.; Simon, M.F.; Gross, E.; Boddey, R.M.; Elliott, G.N.; Neto, N.E.; de Fatima Loureiro, M.; de Queiroz, L.P.; Scotti, M.R.; Chen, W.M.; et al. Nodulation and Nitrogen Fixation by Mimosa spp. in the Cerrado and Caatinga Biomes of Brazil. New Phytol. 2010, 186, 934–946. [Google Scholar] [CrossRef]
- Taulé, C.; Zabaleta, M.; Mareque, C.; Platero, R.; Sanjurjo, L.; Sicardi, M.; Frioni, L.; Battistoni, F.; Fabiano, E. New Betaproteobacterial Rhizobium Strains Able to Efficiently Nodulate Parapiptadenia Rigida (Benth.) Brenan. Appl. Environ. Microbiol. 2012, 78, 1692–1700. [Google Scholar] [CrossRef]
- Dludlu, M.N.; Chimphango, S.B.M.; Walker, G.; Stirton, C.H.; Muasya, A.M. Horizontal Gene Transfer among Rhizobia of the Core Cape Subregion of Southern Africa. S. Afr. J. Bot. 2018, 118, 342–352. [Google Scholar] [CrossRef]
- Beukes, C.W.; Venter, S.N.; Steenkamp, E.T. The History and Distribution of Nodulating Paraburkholderia, a Potential Inoculum for Fynbos Forage Species. Grass Forage Sci. 2021, 76, 10–32. [Google Scholar] [CrossRef]
- de Castro Pires, R.; dos Reis Junior, F.B.; Zilli, J.E.; Fischer, D.; Hofmann, A.; James, E.K.; Simon, M.F. Soil Characteristics Determine the Rhizobia in Association with Different Species of Mimosa in Central Brazil. Plant. Soil. 2018, 423, 411–428. [Google Scholar] [CrossRef]
- Soares Neto, C.B.; Ribeiro, P.R.A.; Fernandes-Júnior, P.I.; de Andrade, L.R.M.; Zilli, J.E.; Mendes, I.C.; do Vale, H.M.M.; James, E.K.; dos Reis Junior, F.B. Paraburkholderia atlantica Is the Main Rhizobial Symbiont of Mimosa spp. in Ultramafic Soils in the Brazilian Cerrado Biome. Plant Soil. 2022, 479, 465–479. [Google Scholar] [CrossRef]
- Cordeiro Silva, V.; Alves Casaes Alves, P.; Ferreira Kruschewsky Rhem, M.; Ferreira dos Santos, J.M.; James, E.K.; Gross, E. Brazilian Species of Calliandra Benth. (Tribe Ingeae) Are Nodulated by Diverse Strains of Paraburkholderia. Syst. Appl. Microbiol. 2018, 41, 241–250. [Google Scholar] [CrossRef]
- Zilli, J.É.; de Moraes Carvalho, C.P.; de Matos Macedo, A.V.; de Barros Soares, L.H.; Gross, E.; James, E.K.; Simon, M.F.; de Faria, S.M. Nodulation of the Neotropical Genus Calliandra by Alpha or Betaproteobacterial Symbionts Depends on the Biogeographical Origins of the Host Species. Braz. J. Microbiol. 2021, 52, 2153–2168. [Google Scholar] [CrossRef]
- Bontemps, C.; Rogel, M.A.; Wiechmann, A.; Mussabekova, A.; Moody, S.; Simon, M.F.; Moulin, L.; Elliott, G.N.; Lacercat-Didier, L.; Dasilva, C.; et al. Endemic Mimosa Species from Mexico Prefer Alphaproteobacterial Rhizobial Symbionts. New Phytol. 2016, 209, 319–333. [Google Scholar] [CrossRef]
- Platero, R.; James, E.K.; Rios, C.; Iriarte, A.; Sandes, L.; Zabaleta, M.; Battistoni, F.; Fabiano, E. Novel Cupriavidus Strains Isolated from Root Nodules of Native Uruguayan Mimosa Species. Appl. Environ. Microbiol. 2016, 82, 3150–3164. [Google Scholar] [CrossRef]
- Schultze, M.; Staehelin, C.; Brunner, F.; Genetet, I.; Legrand, M.; Fritig, B.; Kondorosi, E.; Kondorosi, A. Plant Chitinase/Lysozyme Isoforms Show Distinct Substrate Specificity and Cleavage Site Preference towards Lipochitooligosaccharide Nod Signals. Plant J. 1998, 16, 571–580. [Google Scholar] [CrossRef]
- Perret, X.; Staehelin, C.; Broughton, W.J. Molecular Basis of Symbiotic Promiscuity. Microbiol. Mol. Biol. Rev. 2000, 64, 180–201. [Google Scholar] [CrossRef]
- Chen, W.M.; Moulin, L.; Bontemps, C.; Vandamme, P.; Béna, G.; Boivin-Masson, C. Legume Symbiotic Nitrogen Fixation by β-Proteobacteria Is Widespread in Nature. J. Bacteriol. 2003, 185, 7266–7272. [Google Scholar] [CrossRef]
- de Meyer, S.E.; Briscoe, L.; Martínez-Hidalgo, P.; Agapakis, C.M.; De-Los Santos, P.E.; Seshadri, R.; Reeve, W.; Weinstock, G.; O’Hara, G.; Howieson, J.G.; et al. Symbiotic Burkholderia Species Show Diverse Arrangements of Nif/Fix and Nod Genes and Lack Typical High-Affinity Cytochrome Cbb3 Oxidase Genes. Mol. Plant-Microbe Interact. 2016, 29, 609–619. [Google Scholar] [CrossRef]
- Sheu, S.Y.; Chou, J.H.; Bontemps, C.; Elliott, G.N.; Gross, E.; dos Reis Junior, F.B.; Melkonian, R.; Moulin, L.; James, E.K.; Sprent, J.I.; et al. Burkholderia Diazotrophica sp. Nov., Isolated from Root Nodules of Mimosa spp. Int. J. Syst. Evol. Microbiol. 2013, 63, 435–441. [Google Scholar] [CrossRef]
- Coenye, T.; Henry, D.; Speert, D.P.; Vandamme, P. Burkholderia phenoliruptrix sp. Nov., to Accommodate the 2,4,5-Trichlorophenoxyacetic Acid and Halophenol-Degrading Strain AC1100. Syst. Appl. Microbiol. 2004, 27, 623–627. [Google Scholar] [CrossRef]
- Bournaud, C.; Moulin, L.; Cnockaert, M.; de Faria, S.; Prin, Y.; Severac, D.; Vandamme, P. Paraburkholderia piptadeniae sp. Nov. and Paraburkholderia ribeironis sp. Nov., Two Root-Nodulating Symbiotic Species of Piptadenia gonoacantha in Brazil. Int. J. Syst. Evol. Microbiol. 2017, 67, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; de Farja, S.M.; Chou, J.H.; James, E.K.; Elliott, G.N.; Sprent, J.I.; Bontemps, C.; Young, J.P.W.; Vandamme, P. Burkholderia sabiae sp. Nov., Isolated from Root Nodules of Mimosa caesalpiniifolia. Int. J. Syst. Evol. Microbiol. 2008, 58, 2174–2179. [Google Scholar] [CrossRef] [PubMed]
- Paulitsch, F.; Dall’Agnol, R.F.; Delamuta, J.R.M.; Ribeiro, R.A.; da Silva Batista, J.S.; Hungria, M. Paraburkholderia atlantica sp. Nov. and Paraburkholderia franconis sp. Nov., Two New Nitrogen-Fixing Nodulating Species Isolated from Atlantic Forest Soils in Brazil. Arch. Microbiol. 2020, 202, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Paulitsch, F.; Dall’Agnol, R.F.; Delamuta, J.R.M.; Ribeiro, R.A.; da Silva Batista, J.S.; Hungria, M. Paraburkholderia guartelaensis sp. Nov., a Nitrogen-Fixing Species Isolated from Nodules of Mimosa Gymnas in an Ecotone Considered as a Hotspot of Biodiversity in Brazil. Arch. Microbiol. 2019, 201, 1435–1446. [Google Scholar] [CrossRef]
- Mavima, L.; Beukes, C.W.; Palmer, M.; de Meyer, S.E.; James, E.K.; Maluk, M.; Gross, E.; dos Reis Junior, F.B.; Avontuur, J.R.; Chan, W.Y.; et al. Paraburkholderia youngii sp. Nov. and ‘Paraburkholderia atlantica’—Brazilian and Mexican Mimosa-Associated Rhizobia That Were Previously Known as Paraburkholderia tuberum Sv. Mimosae. Syst. Appl. Microbiol. 2021, 44, 126152. [Google Scholar] [CrossRef]
- de Meyer, S.E.; Cnockaert, M.; Ardley, J.K.; van Wyk, B.E.; Vandamme, P.A.; Howieson, J.G. Burkholderia dilworthii sp. Nov., Isolated from Lebeckia ambigua Root Nodules. Int. J. Syst. Evol. Microbiol. 2014, 64, 1090–1095. [Google Scholar] [CrossRef]
- Steenkamp, E.T.; van Zyl, E.; Beukes, C.W.; Avontuur, J.R.; Chan, W.Y.; Palmer, M.; Mthombeni, L.S.; Phalane, F.L.; Sereme, T.K.; Venter, S.N. Burkholderia kirstenboschensis sp. Nov. Nodulates Papilionoid Legumes Indigenous to South Africa. Syst. Appl. Microbiol. 2015, 38, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Beukes, C.W.; Steenkamp, E.T.; van Zyl, E.; Avontuur, J.; Chan, W.Y.; Hassen, A.I.; Palmer, M.; Mthombeni, L.S.; Phalane, F.L.; Sereme, T.K.; et al. Paraburkholderia strydomiana sp. Nov. and Paraburkholderia steynii sp. Nov.: Rhizobial Symbionts of the Fynbos Legume Hypocalyptus sophoroides. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2019, 112, 1369–1385. [Google Scholar] [CrossRef] [PubMed]
- de Meyer, S.E.; Cnockaert, M.; Ardley, J.K.; Trengove, R.D.; Garau, G.; Howieson, J.G.; Vandamme, P. Burkholderia rhynchosiae sp. Nov., Isolated from Rhynchosia ferulifolia Root Nodules. Int. J. Syst. Evol. Microbiol. 2013, 63, 3944–3949. [Google Scholar] [CrossRef]
- de Meyer, S.E.; Cnockaert, M.; Ardley, J.K.; Maker, G.; Yates, R.; Howieson, J.G.; Vandamme, P. Burkholderia sprentiae sp. Nov., Isolated from Lebeckia ambigua Root Nodules. Int. J. Syst. Evol. Microbiol. 2013, 63, 3950–3957. [Google Scholar] [CrossRef]
- Reeve, W.; de Meyer, S.; Terpolilli, J.; Melino, V.; Ardley, J.; Rui, T.; Tiwari, R.; Howieson, J.; Yates, R.; O’Hara, G. Genome Sequence of the Lebeckia Ambigua-Nodulating “Burkholderia sprentiae” Strain WSM5005T. Stand. Genom. Sci. 2013, 9, 385–394. [Google Scholar] [CrossRef][Green Version]
- Rogel, M.A.; Ormeño-Orrillo, E.; Martinez Romero, E. Symbiovars in Rhizobia Reflect Bacterial Adaptation to Legumes. Syst. Appl. Microbiol. 2011, 34, 96–104. [Google Scholar] [CrossRef]
- Andrews, M.; de Meyer, S.; James, E.K.; Stępkowski, T.; Hodge, S.; Simon, M.F.; Young, J.P.W. Horizontal Transfer of Symbiosis Genes within and between Rhizobial Genera: Occurrence and Importance. Genes 2018, 9, 321. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.; Hawkins, J.A. Are Cape Floral Clades the Same Age? Contemporaneous Origins of Two Lineages in the Genistoids s.l. (Fabaceae). Mol. Phylogenet. Evol. 2007, 45, 952–970. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.F.; Grether, R.; de Queiroz, L.P.; Skema, C.; Pennington, R.T.; Hughes, C.E. Recent Assembly of the Cerrado, a Neotropical Plant Diversity Hotspot, by in Situ Evolution of Adaptations to Fire. Proc. Natl. Acad. Sci. USA 2009, 106, 20359–20364. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.; Herendeen, P.S.; Wojciechowski, M.F. Evolutionary Rates Analysis of Leguminosae Implicates a Rapid Diversification of Lineages during the Tertiary. Syst. Biol. 2005, 54, 575–594. [Google Scholar] [CrossRef]
- Bauer, W.D.; Caetano-Anollés, G. Chemotaxis, Induced Gene Expression and Competitiveness in the Rhizosphere. Plant Soil 1990, 129, 45–52. [Google Scholar] [CrossRef]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within Symbiosis: Evolving Nitrogen-Fixing Legume Symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef]
- Atieno, M.; Lesueur, D. Opportunities for Improved Legume Inoculants: Enhanced Stress Tolerance of Rhizobia and Benefits to Agroecosystems. Symbiosis 2019, 77, 191–205. [Google Scholar] [CrossRef]
- Basile, L.A.; Lepek, V.C. Legume–Rhizobium Dance: An Agricultural Tool That Could Be Improved? Microb. Biotechnol. 2021, 14, 1897–1917. [Google Scholar] [CrossRef]
- Hug, S.; Liu, Y.; Heiniger, B.; Bailly, A.; Ahrens, C.H.; Eberl, L.; Pessi, G. Differential Expression of Paraburkholderia phymatum Type VI Secretion Systems (T6SS) Suggests a Role of T6SS-b in Early Symbiotic Interaction. Front. Plant Sci. 2021, 12, 699590. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, A.; Oliveira, S. Response to Temperature Stress in Rhizobia. Crit. Rev. Microbiol. 2013, 39, 219–228. [Google Scholar] [CrossRef]
- Goyal, R.K.; Mattoo, A.K.; Schmidt, M.A. Rhizobial–Host Interactions and Symbiotic Nitrogen Fixation in Legume Crops toward Agriculture Sustainability. Front. Microbiol. 2021, 12, 669404. [Google Scholar] [CrossRef]
- Estévez, J.; Soria-Díaz, M.E.; de Córdoba, F.F.; Morón, B.; Manyani, H.; Gil, A.; Thomas-Oates, J.; van Brussel, A.A.N.; Dardanelli, M.S.; Sousa, C.; et al. Different and New Nod Factors Produced by Rhizobium tropici CIAT899 Following Na+ Stress. FEMS Microbiol. Lett. 2009, 293, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Talbi, C.; Argandoña, M.; Salvador, M.; Alché, J.D.; Vargas, C.; Bedmar, E.J.; Delgado, M.J. Burkholderia phymatum Improves Salt Tolerance of Symbiotic Nitrogen Fixation in Phaseolus vulgaris. Plant Soil. 2013, 367, 673–685. [Google Scholar] [CrossRef]
- Ferreira, A.S.; Leitão, J.H.; Silva, I.N.; Pinheiros, P.F.; Sousa, S.A.; Ramos, C.G.; Moreira, L.M. Distribution of Cepacian Biosynthesis Genes among Environmental and Clinical Burkholderia Strains and Role of Cepacian Exopolysaccharide in Resistance to Stress Conditions. Appl. Environ. Microbiol. 2010, 76, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bellich, B.; Hug, S.; Eberl, L.; Cescutti, P.; Pessi, G. The Exopolysaccharide Cepacian Plays a Role in the Establishment of the Paraburkholderia phymatum—Phaseolus vulgaris Symbiosis. Front. Microbiol. 2020, 11, 1600. [Google Scholar] [CrossRef]
- Ferreira, A.S.; Silva, I.N.; Oliveira, V.H.; Cunha, R.; Moreira, L.M. Insights into the Role of Extracellular Polysaccharides in Burkholderia Adaptation to Different Environments. Front. Cell. Infect. Microbiol. 2011, 1, 16. [Google Scholar] [CrossRef]
- Lagares, A.; Draghi, W.O.; del Papa, M.F.; Hellweg, C.; Watt, S.A.; Watt, T.F.; Barsch, A.; Lozano, M.J.; Lagares, A.; Salas, M.E.; et al. A Consolidated Analysis of the Physiologic and Molecular Responses Induced under Acid Stress in the Legume-Symbiont Model-Soil Bacterium Sinorhizobium meliloti. Sci. Rep. 2016, 6, 29278. [Google Scholar] [CrossRef]
- Stopnisek, N.; Bodenhausen, N.; Frey, B.; Fierer, N.; Eberl, L.; Weisskopf, L. Genus-wide Acid Tolerance Accounts for the Biogeographical Distribution of Soil Burkholderia Populations. Environ. Microbiol. 2014, 16, 1503–1512. [Google Scholar] [CrossRef]
- Weisskopf, L.; Heller, S.; Eberl, L. Burkholderia Species Are Major Inhabitants of White Lupin Cluster Roots. Appl. Environ. Microbiol. 2011, 77, 7715–7720. [Google Scholar] [CrossRef] [PubMed]
- Kost, T.; Stopnisek, N.; Agnoli, K.; Eberl, L.; Weisskopf, L. Oxalotrophy, a Widespread Trait of Plant-Associated Burkholderia Species, Is Involved in Successful Root Colonization of Lupin and Maize by Burkholderia phytofirmans. Front. Microbiol. 2014, 4, 421. [Google Scholar] [CrossRef] [PubMed]
- Giller, K.E.; Witter, E.; Mcgrath, S.P. Toxicity of Heavy Metals to Microorganisms and Microbial Processes in Agricultural Soils: A Review. Soil Biol. Biochem. 1998, 30, 1389–1414. [Google Scholar] [CrossRef]
- Poole, P. Shining a Light on the Dark World of Plant Root-Microbe Interactions. Proc. Natl. Acad. Sci. USA 2017, 114, 4281–4283. [Google Scholar] [CrossRef] [PubMed]
- Stubbendieck, R.M.; Straight, P.D. Multifaceted Interfaces of Bacterial Competition. J. Bacteriol. 2016, 198, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Checcucci, A.; DiCenzo, G.C.; Bazzicalupo, M.; Mengoni, A. Trade, Diplomacy, and Warfare: The Quest for Elite Rhizobia Inoculant Strains. Front. Microbiol. 2017, 8, 02207. [Google Scholar] [CrossRef]
- Fields, B.; Moffat, E.K.; Harrison, E.; Andersen, S.U.; Young, J.P.W.; Friman, V.P. Genetic Variation Is Associated with Differences in Facilitative and Competitive Interactions in the Rhizobium leguminosarum Species Complex. Environ. Microbiol. 2022, 24, 3463–3485. [Google Scholar] [CrossRef]
- Onishchuk, O.P.; Vorobyov, N.I.; Provorov, N.A. Nodulation Competitiveness of Nodule Bacteria: Genetic Control and Adaptive Significance. Appl. Biochem. Microbiol. 2017, 53, 131–139. [Google Scholar] [CrossRef]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A.H.M. Impact of Root Exudates and Plant Defense Signaling on Bacterial Communities in the Rhizosphere: A Review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- Lardi, M.; de Campos, S.B.; Purtschert, G.; Eberl, L.; Pessi, G. Competition Experiments for Legume Infection Identify Burkholderia phymatum as a Highly Competitive β-Rhizobium. Front. Microbiol. 2017, 8, 01527. [Google Scholar] [CrossRef]
- Kraepiel, A.M.L.; Bellenger, J.P.; Wichard, T.; Morel, F.M.M. Multiple Roles of Siderophores in Free-Living Nitrogen-Fixing Bacteria. BioMetals 2009, 22, 573–581. [Google Scholar] [CrossRef] [PubMed]
- diCenzo, G.C.; MacLean, A.M.; Milunovic, B.; Golding, G.B.; Finan, T.M. Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction. PLoS Genet. 2014, 10, 1004742. [Google Scholar] [CrossRef] [PubMed]
- Klonowska, A.; Melkonian, R.; Miché, L.; Tisseyre, P.; Moulin, L. Transcriptomic Profiling of Burkholderia phymatum STM815, Cupriavidus taiwanensis LMG19424 and Rhizobium mesoamericanum STM3625 in Response to Mimosa pudica Root Exudates Illuminates the Molecular Basis of Their Nodulation Competitiveness and Symbiotic Evolutionary History. BMC Genom. 2018, 19, 1–22. [Google Scholar] [CrossRef]
- Zatakia, H.M.; Nelson, C.E.; Syed, U.J.; Scharf, B.E. ExpR Coordinates the Expression of Symbiotically Important, Bundle-Forming Flp Pili with Quorum Sensing in Sinorhizobium meliloti. Appl. Environ. Microbiol. 2014, 80, 2429–2439. [Google Scholar] [CrossRef] [PubMed]
- Yost, C.K.; Rochepeau, P.; Hynes, M.F. Rhizobium leguminosarum Contains a Group of Genes That Appear to Code for Methyl-Accepting Chemotaxis Proteins. Microbiology 1998, 144, 1945–1946. [Google Scholar] [CrossRef]
- Lardi, M.; Liu, Y.; Hug, S.; Bolzan de Campos, S.; Eberl, L.; Pessi, G. Paraburkholderia phymatum STM815 σ54 Controls Utilization of Dicarboxylates, Motility, and T6SS-b Expression. Nitrogen 2020, 1, 81–98. [Google Scholar] [CrossRef]
- Downie, J.A. The Roles of Extracellular Proteins, Polysaccharides and Signals in the Interactions of Rhizobia with Legume Roots. FEMS Microbiol. Rev. 2010, 34, 150–170. [Google Scholar] [CrossRef]
- Coutinho, B.G.; Mitter, B.; Talbi, C.; Sessitsch, A.; Bedmar, E.J.; Halliday, N.; James, E.K.; Cámara, M.; Venturi, V. Regulon Studies and in Planta Role of the BraI/R Quorum-Sensing System in the Plant-Beneficial Burkholderia Cluster. Appl. Environ. Microbiol. 2013, 79, 4421–4432. [Google Scholar] [CrossRef]
- Zacharof, M.P.; Lovitt, R.W. Bacteriocins Produced by Lactic Acid Bacteria. APCBEE Procedia 2012, 2, 50–56. [Google Scholar] [CrossRef]
- Cleveland, J.; Montville, T.J.; Nes, I.F.; Chikindas, M.L. Bacteriocins: Safe, Natural Antimicrobials for Food Preservation. Int. J. Food Microbiol. 2001, 71, 1–20. [Google Scholar] [CrossRef]
- Hafeez, F.Y.; Naeem, F.I.; Naeem, R.; Zaidi, A.H.; Malik, K.A. Symbiotic Effectiveness and Bacteriocin Production by Rhizobium Leguminosarum Bv. Viciae Isolated from Agriculture Soils in Faisalabad. Environ. Exp. Bot. 2005, 54, 142–147. [Google Scholar] [CrossRef]
- Maan, P.K.; Garcha, S. Bacteriocins from Gram-Negative Rhizobium spp. Artic. Adv. Bioresearch 2018, 9, 3643. [Google Scholar] [CrossRef]
- Tapia-García, E.Y.; Hernández-Trejo, V.; Guevara-Luna, J.; Rojas-Rojas, F.U.; Arroyo-Herrera, I.; Meza-Radilla, G.; Vásquez-Murrieta, M.S.; Estrada-de los Santos, P. Plant Growth-Promoting Bacteria Isolated from Wild Legume Nodules and Nodules of Phaseolus vulgaris L. Trap Plants in Central and Southern Mexico. Microbiol. Res. 2020, 239, 126522. [Google Scholar] [CrossRef]
- Wang, K.; Wu, Y.; Ye, M.; Yang, Y.; Asiegbu, F.O.; Overmyer, K.; Liu, S.; Cui, F. Comparative Genomics Reveals Potential Mechanisms of Plant Beneficial Effects of a Novel Bamboo-Endophytic Bacterial Isolate Paraburkholderia sacchari Suichang626. Front. Microbiol. 2021, 12, 686988. [Google Scholar] [CrossRef] [PubMed]
- Bach, E.; Passaglia, L.M.P.; Jiao, J.; Gross, H. Burkholderia in the Genomic Era: From Taxonomy to the Discovery of New Antimicrobial Secondary Metabolites. Crit. Rev. Microbiol. 2021, 48, 121–160. [Google Scholar] [CrossRef] [PubMed]
- Horsman, M.E.; Marous, D.R.; Li, R.; Oliver, R.A.; Byun, B.; Emrich, S.J.; Boggess, B.; Townsend, C.A.; Mobashery, S. Whole-Genome Shotgun Sequencing of Two β-Proteobacterial Species in Search of the Bulgecin Biosynthetic Cluster. ACS Chem. Biol. 2017, 12, 2552–2557. [Google Scholar] [CrossRef]
- Tomoshige, S.; Dik, D.A.; Akabane-Nakata, M.; Madukoma, C.S.; Fisher, J.F.; Shrout, J.D.; Mobashery, S. Total Syntheses of Bulgecins A, B, and C and Their Bactericidal Potentiation of the β-Lactam Antibiotics. ACS Infect. Dis. 2018, 4, 860–867. [Google Scholar] [CrossRef]
- Aoki, S.K.; Diner, E.J.; de Roodenbeke, C.T.K.; Burgess, B.R.; Poole, S.J.; Braaten, B.A.; Jones, A.M.; Webb, J.S.; Hayes, C.S.; Cotter, P.A.; et al. A Widespread Family of Polymorphic Contact-Dependent Toxin Delivery Systems in Bacteria. Nature 2010, 468, 439–442. [Google Scholar] [CrossRef]
- Anderson, M.S.; Garcia, E.C.; Cotter, P.A. The Burkholderia BcpAIOB Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems. PLoS Genet. 2012, 8, 1002877. [Google Scholar] [CrossRef]
- Meuskens, I.; Saragliadis, A.; Leo, J.C.; Linke, D. Type V Secretion Systems: An Overview of Passenger Domain Functions. Front. Microbiol. 2019, 10, 01163. [Google Scholar] [CrossRef] [PubMed]
- Ikryannikova, L.N.; Kurbatov, L.K.; Gorokhovets, N.V.; Zamyatnin, A.A. Contact-Dependent Growth Inhibition in Bacteria: Do Not Get Too Close! Int. J. Mol. Sci. 2020, 21, 7990. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.C.; Perault, A.I.; Marlatt, S.A.; Cotter, P.A. Interbacterial Signaling via Burkholderia Contact Dependent Growth Inhibition System Proteins. Proc. Natl. Acad. Sci. USA 2016, 113, 8296–8301. [Google Scholar] [CrossRef] [PubMed]
- Perault, A.I.; Cotter, P.A. Three Distinct Contact-Dependent Growth Inhibition Systems Mediate Interbacterial Competition by the Cystic Fibrosis Pathogen Burkholderia dolosa. J. Bacteriol. 2018, 200, e00428-18. [Google Scholar] [CrossRef]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, Load, Fire: The Type VI Secretion System, a Bacterial Nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef]
- Roest, H.P.; Mulders, I.H.M.; Spaink, H.P.; Wijffelman, C.A.; Lugtenberg, B.J.J. A Rhizobium leguminosarum Biovar Trifolii Locus Not Localized on the Sym Plasmid Hinders Effective Nodulation on Plants of the Pea Cross-Inoculation Group. Mol. Plant. Microbe Interact. 1997, 10, 938–941. [Google Scholar] [CrossRef]
- Bladergroen, M.R.; Badelt, K.; Spaink, H.P. Infection-Blocking Genes of a Symbiotic Rhizobium Leguminosarum Strain That Are Involved in Temperature-Dependent Protein Secretion. Mol. Plant. Microbe Interact. 2003, 16, 53–64. [Google Scholar] [CrossRef]
- Ho, B.T.; Dong, T.G.; Mekalanos, J.J. A View to a Kill: The Bacterial Type VI Secretion System. Cell Host Microbe 2014, 15, 9–21. [Google Scholar] [CrossRef]
- de Campos, S.B.; Lardi, M.; Gandolfi, A.; Eberl, L.; Pessi, G. Mutations in Two Paraburkholderia phymatum Type VI Secretion Systems Cause Reduced Fitness in Interbacterial Competition. Front. Microbiol. 2017, 8, 2473. [Google Scholar] [CrossRef]
- Gage, D.J. Infection and Invasion of Roots by Symbiotic, Nitrogen-Fixing Rhizobia during Nodulation of Temperate Legumes. Microbiol. Mol. Biol. Rev. 2004, 68, 280–300. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D. Speak, Friend, and Enter: Signalling Systems That Promote Beneficial Symbiotic Associations in Plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Udvardi, M.; Poole, P.S. Transport and Metabolism in Legume-Rhizobia Symbioses. Annu. Rev. Plant. Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef]
- Hassen, A.I.; Lamprecht, S.C.; Bopape, F.L. Emergence of β-Rhizobia as New Root Nodulating Bacteria in Legumes and Current Status of the Legume–Rhizobium Host Specificity Dogma. World J. Microbiol. Biotechnol. 2020, 36. [Google Scholar] [CrossRef]
- Okubo, T.; Fukushima, S.; Minamisawa, K. Evolution of Bradyrhizobium-Aeschynomene Mutualism: Living Testimony of the Ancient World or Highly Evolved State? Plant. Cell Physiol. 2012, 53, 2000–2007. [Google Scholar] [CrossRef]
- Salinero-Lanzarote, A.; Pacheco-Moreno, A.; Domingo-Serrano, L.; Durán, D.; Ormeño-Orrillo, E.; Martínez-Romero, E.; Albareda, M.; Palacios, J.M.; Rey, L. The Type VI Secretion System of Rhizobium etli Mim1 Has a Positive Effect in Symbiosis. FEMS Microbiol. Ecol. 2019, 95, fiz054. [Google Scholar] [CrossRef] [PubMed]
- Dénarié, J.; Debellé, F.; Promé, J.-C. Rhizobium Lipo-Chitooligosaccharide Nodulation Factors: Signaling Molecules Mediating Recognition and Morphogenesis. Annu. Rev. Biochem. 1996, 65, 503–553. [Google Scholar] [CrossRef]
- D’haeze, W.; Holsters, M. Nod Factor Structures, Responses, and Perception during Initiation of Nodule Development. Glycobiology 2002, 12, 79R–105R. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.; Maillet, F.; Plazanet, C.; Debellé, F.; Ferro, M.; Truchet, G.; Promé, J.-C.; Dénarié, J. The Common NodABC Genes of Rhizobium meliloti Are Host-Range Determinants. Proc. Natl. Acad. Sci. USA 1996, 93, 15305–15310. [Google Scholar] [CrossRef] [PubMed]
- Mergaert, P.; van Montagu, M.; Holsters, M. Molecular Mechanisms of Nod Factor Diversity. Mol. Microbiol. 1997, 25, 811–817. [Google Scholar] [CrossRef]
- Ghantasala, S.; Roy Choudhury, S. Nod Factor Perception: An Integrative View of Molecular Communication during Legume Symbiosis. Plant. Mol. Biol. 2022, 110, 485–509. [Google Scholar] [CrossRef]
- Martínez-Romero, E. Coevolution in Rhizobium-Legume Symbiosis? DNA Cell Biol. 2009, 28, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Geddes, B.A.; Kearsley, J.; Morton, R.; diCenzo, G.C.; Finan, T.M. The Genomes of Rhizobia. In Advances in Botanical Research; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 94, pp. 213–249. ISBN 9780081027981. [Google Scholar]
- Daubech, B.; Poinsot, V.; Klonowska, A.; Capela, D.; Chaintreuil, C.; Moulin, L.; Marchetti, M.; Masson-Boivin, C. NoeM, a New Nodulation Gene Involved in the Biosynthesis of Nod Factors with an Open-Chain Oxidized Terminal Residue and in the Symbiosis with Mimosa Pudica. Mol. Plant.-Microbe Interact. 2019, 32, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Hirsch, A.M. Signals and Responses: Choreographing the Complex Interaction between Legumes and α- and β-Rhizobia. Plant. Signal. Behav. 2006, 1, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Rinaudi, L.V.; Giordano, W. An Integrated View of Biofilm Formation in Rhizobia. FEMS Microbiol. Lett. 2010, 304, 1–11. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From Saprophytes to Endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS Matrix: The “House of Biofilm Cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef]
- Flemming, H.; Wingender, J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Via, V.D.; Zanetti, M.E.; Blanco, F. How Legumes Recognize Rhizobia. Plant. Signal. Behav. 2016, 11, 1–6. [Google Scholar] [CrossRef]
- Clúa, J.; Roda, C.; Zanetti, M.E.; Blanco, F.A. Compatibility between Legumes and Rhizobia for the Establishment of a Successful Nitrogen-Fixing Symbiosis. Genes 2018, 9, 125. [Google Scholar] [CrossRef]
- Vallenet, D.; Engelen, S.; Mornico, D.; Cruveiller, S.; Fleury, L.; Lajus, A.; Rouy, Z.; Roche, D.; Salvignol, G.; Scarpelli, C.; et al. MicroScope: A Platform for Microbial Genome Annotation and Comparative Genomics. Database 2009, 2009, bap021. [Google Scholar] [CrossRef] [PubMed]
- Crespi, M.; Frugier, F. De Novo Organ Formation from Differentiated Cells: Root Nodule Organogenesis. Dev. Biol. 2008, 1, re11. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Indrasumunar, A.; Hayashi, S.; Lin, M.H.; Lin, Y.H.; Reid, D.E.; Gresshoff, P.M. Molecular Analysis of Legume Nodule Development and Autoregulation. J. Integr. Plant. Biol. 2010, 52, 61–76. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin Biosynthesis and Its Role in Plant Development. Annu. Rev. Plant. Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, C.; Yang, J.; Yu, N.; Wang, E. Hormone Modulation of Legume-Rhizobial Symbiosis. J. Integr. Plant. Biol. 2018, 60, 632–648. [Google Scholar] [CrossRef]
- Went, F.W.; Thimann, K.V. Phytohormones; The Macmillan Company: New York, NY, USA, 1937. [Google Scholar]
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-Acetic Acid in Plant-Microbe Interactions. Antonie Leeuwenhoek 2014, 106, 85–125. [Google Scholar] [CrossRef]
- Bellés-Sancho, P.; Liu, Y.; Heiniger, B.; von Salis, E.; Eberl, L.; Ahrens, C.H.; Zamboni, N.; Bailly, A.; Pessi, G. A Novel Function of the Key Nitrogen-Fixation Activator NifA in Beta-Rhizobia: Repression of Bacterial Auxin Synthesis during Symbiosis. Front. Plant. Sci. 2022, 13, 991548. [Google Scholar] [CrossRef]
- Lardi, M.; Liu, Y.; Purtschert, G.; de Campos, S.B.; Pessi, G. Transcriptome Analysis of Paraburkholderia phymatum under Nitrogen Starvation and during Symbiosis with Phaseolus vulgaris. Genes 2017, 8, 8120389. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, M.; Okazaki, S.; Nukui, N.; Ezura, H.; Mitsui, H.; Minamisawa, K. Rhizobitoxine Modulates Plant-Microbe Interactions by Ethylene Inhibition. Biotechnol. Adv. 2006, 24, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Tavares, M.J.; Rossi, J.; Glick, B.R. The Modulation of Leguminous Plant Ethylene Levels by Symbiotic Rhizobia Played a Role in the Evolution of the Nodulation Process. Heliyon 2018, 4, 1068. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; Prell, J.; James, E.K.; Sheu, D.S.; Sheu, S.Y. Effect of Phosphoglycerate Mutase and Fructose 1,6-Bisphosphatase Deficiency on Symbiotic Burkholderia phymatum. Microbiology 2012, 158, 1127–1136. [Google Scholar] [CrossRef]
- Clarke, V.C.; Loughlin, P.C.; Day, D.A.; Smith, P.M.C. Transport Processes of the Legume Symbiosome Membrane. Front. Plant. Sci. 2014, 5, 699. [Google Scholar] [CrossRef]
- Poole, P.S.; Ledermann, R. Maintaining Osmotic Balance in Legume Nodules. J. Exp. Bot. 2022, 73, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.M.; Ludden, P.W. Maturation of Nitrogenase: A Biochemical Puzzle. J. Bacteriol. 2005, 187, 405–414. [Google Scholar] [CrossRef]
- Fischer, H.M. Genetic Regulation of Nitrogen Fixation in Rhizobia. Microbiol. Rev. 1994, 58, 352–386. [Google Scholar] [CrossRef]
- Hauser, F.; Pessi, G.; Friberg, M.; Weber, C.; Rusca, N.; Lindemann, A.; Fischer, H.M.; Hennecke, H. Dissection of the Bradyrhizobium japonicum NifA+σ54 Regulon, and Identification of a Ferredoxin Gene (FdxN) for Symbiotic Nitrogen Fixation. Mol. Genet. Genom. 2007, 278, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Lardi, M.; Liu, Y.; Giudice, G.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomics and Transcriptomics Identify Multiple Downstream Targets of Paraburkholderia phymatum σ54 during Symbiosis with Phaseolus vulgaris. Int. J. Mol. Sci. 2018, 19, 1049. [Google Scholar] [CrossRef] [PubMed]
- Bellés-Sancho, P.; Lardi, M.; Liu, Y.; Eberl, L.; Zamboni, N.; Bailly, A.; Pessi, G. Metabolomics and Dual RNA-sequencing on Root Nodules Revealed New Cellular Functions Controlled by Paraburkholderia phymatum NifA. Metabolites 2021, 11, 455. [Google Scholar] [CrossRef] [PubMed]
- Webb, I.U.C.; Xu, J.; Sanchez-Cañizares, C.; Karunakaran, R.; Ramachandran, V.K.; Rutten, P.J.; East, A.K.; Huang, W.E.; Watmough, N.J.; Poole, P.S. Regulation and Characterization of Mutants of FixABCX in Rhizobium leguminosarum. Mol. Plant.-Microbe Interact. 2021, 34, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Lery, L.M.S.; Bitar, M.; Costa, M.G.S.; Rössle, S.C.S.; Bisch, P.M. Unraveling the Molecular Mechanisms of Nitrogenase Conformational Protection against Oxygen in Diazotrophic Bacteria. BMC Genom. 2010, 11, S7. [Google Scholar] [CrossRef]
- Nouwen, N.; Arrighi, J.F.; Cartieaux, F.; Chaintreuil, C.; Gully, D.; Klopp, C.; Giraud, E. The Role of Rhizobial (NifV) and Plant (FEN1) Homocitrate Synthases in Aeschynomene/Photosynthetic Bradyrhizobium Symbiosis. Sci. Rep. 2017, 7, 448. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Wongdee, J.; Songwattana, P.; Greetatorn, T.; Goto, K.; Tittabutr, P.; Boonkerd, N.; Teaumroong, N.; Uchiumi, T. Homocitrate Synthase Genes of Two Wide-Host-Range Bradyrhizobium Strains Are Differently Required for Symbiosis Depending on Host Plants. Microbes Environ. 2019, 34, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Hakoyama, T.; Niimi, K.; Watanabe, H.; Tabata, R.; Matsubara, J.; Sato, S.; Nakamura, Y.; Tabata, S.; Jichun, L.; Matsumoto, T.; et al. Host Plant Genome Overcomes the Lack of a Bacterial Gene for Symbiotic Nitrogen Fixation. Nature 2009, 462, 514–517. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, R.; Schulte, C.C.M.; Poole, P.S. How Rhizobia Adapt to the Nodule Environment. J. Bacteriol. 2021, 203, e00539-20. [Google Scholar] [CrossRef] [PubMed]
- Yurgel, S.N.; Kahn, M.L. Sinorhizobium Meliloti DctA Mutants with Partial Ability to Transport Dicarboxylic Acids. J. Bacteriol. 2005, 187, 1161–1172. [Google Scholar] [CrossRef]
- Gourion, B.; Delmotte, N.; Bonaldi, K.; Nouwen, N.; Vorholt, J.A.; Giraud, E. Bacterial RuBisCO Is Required for Efficient Bradyrhizobium/Aeschynomene Symbiosis. PLoS ONE 2011, 6, 1900. [Google Scholar] [CrossRef]
- Torres, A.R.; Brito, B.; Imperial, J.; Palacios, J.M.; Ciampitti, I.A.; Ruiz-Argüeso, T.; Hungria, M. Hydrogen-Uptake Genes Improve Symbiotic Efficiency in Common Beans (Phaseolus vulgaris L.). Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2020, 113, 687–696. [Google Scholar] [CrossRef]
- Koch, M.; Delmotte, N.; Rehrauer, H.; Vorholt, J.A.; Pessi, G.; Hennecke, H. Rhizobial Adaptation to Hosts, a New Facet in the Legume Root-Nodule Symbiosis. Mol. Plant.-Microbe Interact. 2010, 23, 784–790. [Google Scholar] [CrossRef]
- Lardi, M.; Murset, V.; Fischer, H.M.; Mesa, S.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures. Int. J. Mol. Sci. 2016, 17, 815. [Google Scholar] [CrossRef]
- Bellés-Sancho, P.; Lardi, M.; Liu, Y.; Hug, S.; Pinto-Carbó, M.A.; Zamboni, N.; Pessi, G. Paraburkholderia phymatum Homocitrate Synthase NifV Plays a Key Role for Nitrogenase Activity during Symbiosis with Papilionoids and in Free-Living Growth Conditions. Cells 2021, 10, 952. [Google Scholar] [CrossRef]
| Gene | Protein Encoded | Functions |
|---|---|---|
| nodA | Acyl transferase | Transfer of fatty acyl chain to the NFs’ backbone |
| nodB | Deacetylase | Removal of acetyl moiety from nitrogen attached to monomer at non-reducing end |
| nodC | NAG transferase | Catenation of monomeric NAG |
| nodD | LysR family transcriptional regulator | Activation of nod genes transcription |
| nodH | Sulphotransferase | Transfers PAPS to reducing end of NFs |
| nodU, nolO | Carbamoyl transferase | Carbamoylation |
| nodI | NF export ATP-binding protein I | Secretion of NFs |
| nodJ | Transport permease protein NodJ | Secretion of NFs |
| nodQ | Subunits of ATP sulfurylase and kinase | Produces PAPS and activates sulphate compounds |
| nodS | Methyltransferase | Addition of methyl group |
| nodT | Outer-membrane lipoprotein | Interacting with NodI and NodJ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellés-Sancho, P.; Beukes, C.; James, E.K.; Pessi, G. Nitrogen-Fixing Symbiotic Paraburkholderia Species: Current Knowledge and Future Perspectives. Nitrogen 2023, 4, 135-158. https://doi.org/10.3390/nitrogen4010010
Bellés-Sancho P, Beukes C, James EK, Pessi G. Nitrogen-Fixing Symbiotic Paraburkholderia Species: Current Knowledge and Future Perspectives. Nitrogen. 2023; 4(1):135-158. https://doi.org/10.3390/nitrogen4010010
Chicago/Turabian StyleBellés-Sancho, Paula, Chrizelle Beukes, Euan K. James, and Gabriella Pessi. 2023. "Nitrogen-Fixing Symbiotic Paraburkholderia Species: Current Knowledge and Future Perspectives" Nitrogen 4, no. 1: 135-158. https://doi.org/10.3390/nitrogen4010010
APA StyleBellés-Sancho, P., Beukes, C., James, E. K., & Pessi, G. (2023). Nitrogen-Fixing Symbiotic Paraburkholderia Species: Current Knowledge and Future Perspectives. Nitrogen, 4(1), 135-158. https://doi.org/10.3390/nitrogen4010010








