Fractional Calculus in Epigenetics: Modelling DNA Methylation Dynamics Using Mittag–Leffler Function
Abstract
1. Introduction
2. Modelling Methylation Dynamics—DNAm vs. Chronological Age
2.1. Standard Exponential Model of DNAm Dynamics
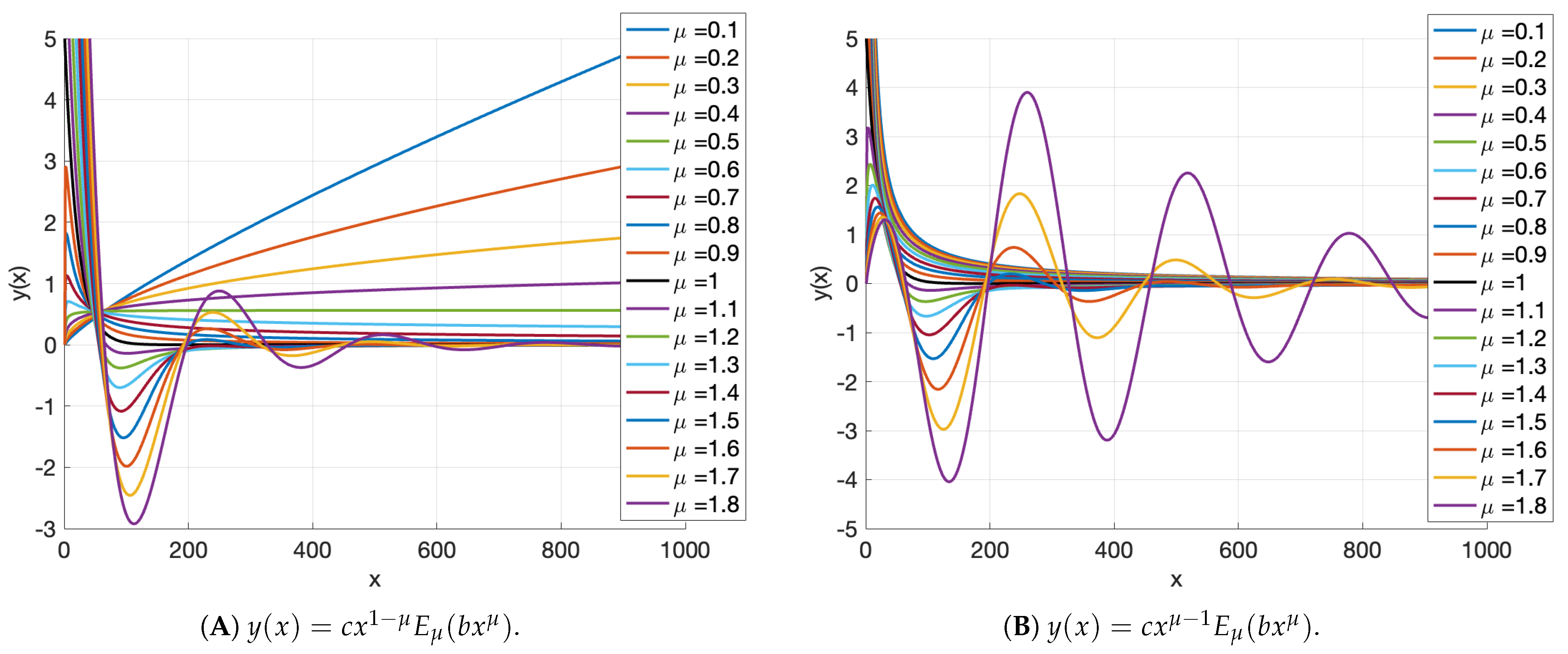
2.2. Fractional Calculus Approach to DNAm Dynamics Modelling
3. Experiments and Discussion on the Results
3.1. DNAm Dataset
3.2. Comparison Metrics for Modelling Performance Evaluation
- Sum of Absolute Errors (SAE) quantifies the total difference between predicted and actual values by summing the absolute value of the discrepancies, serving as a measure of the overall deviation in a dataset.
- Sum of Squared Errors (SSE) measures the unexplained variance in the dependent variable that the model does not account for, with the primary aim to minimise the SSE value, resulting in a model that better fits the data.
- Mean Absolute Percentage Error (MAPE) measures the average percentage difference between predicted and actual values, indicating forecast accuracy by averaging the absolute percentage errors across a set of data points, with a lower value signifying a more accurate model.
- R-squared () as a coefficient of determination from the interval , indicates the proportion of the variance in the dependent variable that can be predicted using the independent variable (one or more), with values closer to one signifying a better fit.
- Adjusted R-squared () accounts for the number of prediction parameters and sample size, penalising the addition of irrelevant independent variables and preventing model overfitting, being a superior criterion for comparing models with different numbers of parameters, providing a more realistic measure of their performance.
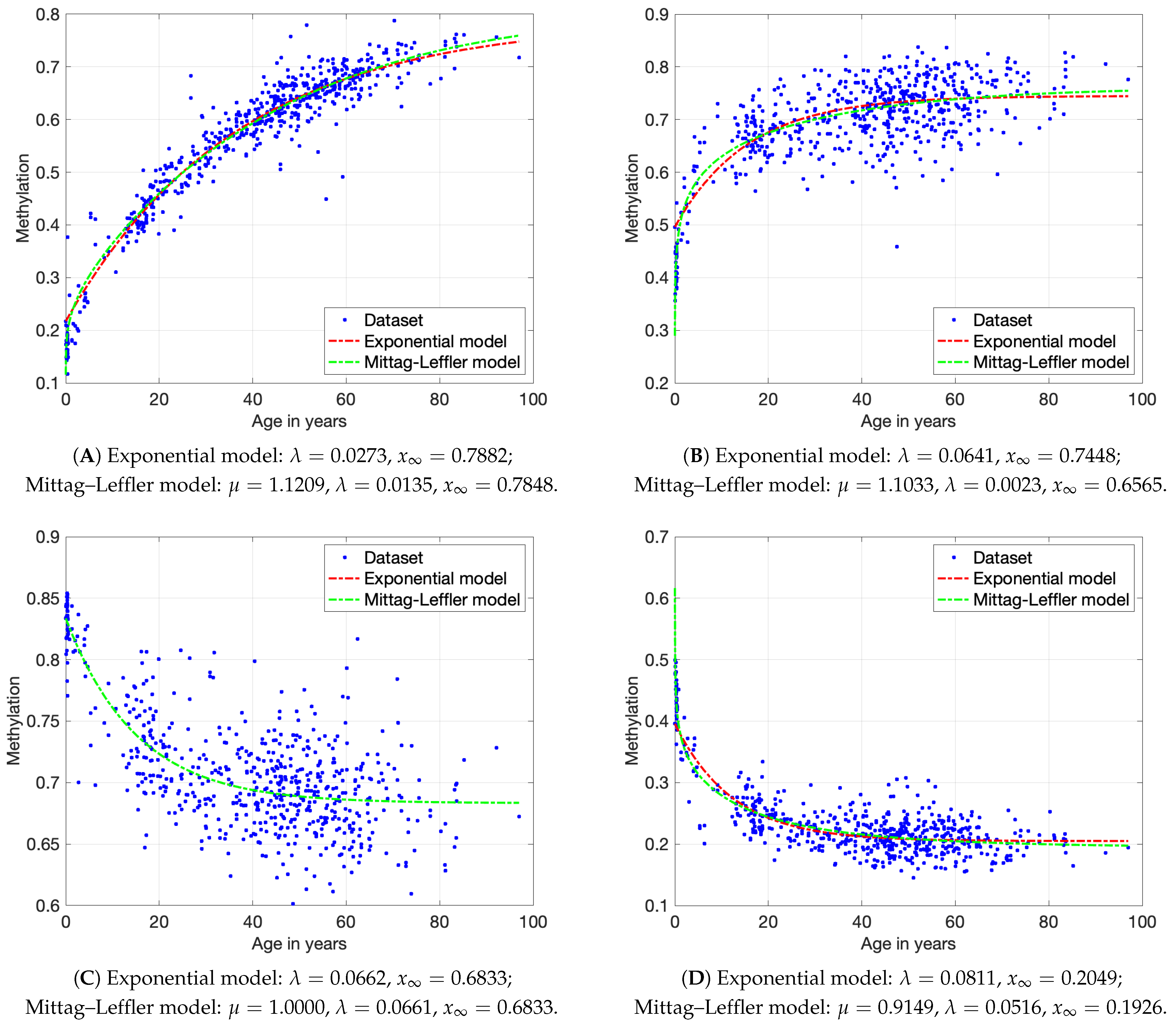
3.3. Mathematical Models of DNAm Dynamics
3.4. Experimental Results
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Mittag–Leffler Function
References
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Cortini, R.; Barbi, M.; Caré, B.R.; Lavelle, C.; Lesne, A.; Mozziconacci, J.; Victor, J.M. The physics of epigenetics. Rev. Mod. Phys. 2016, 88, 025002. [Google Scholar] [CrossRef]
- Guillemin, A.; Stumpf, M.P.H. Non-equilibrium statistical physics, transitory epigenetic landscapes, and cell fate decision dynamics. Math. Biosci. Eng. 2020, 17, 7916–7930. [Google Scholar] [CrossRef]
- Bocklandt, S.; Lin, W.; Sehl, M.E.; Sánchez, F.J.; Sinsheimer, J.S.; Horvath, S.; Vilain, E. Epigenetic predictor of age. PLoS ONE 2011, 6, e14821. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, B.H.; Marioni, R.E.; Colicino, E.; Peters, M.J.; Ward-Caviness, C.K.; Tsai, P.C.; Roetker, N.S.; Just, A.C.; Demerath, E.W.; Guan, W.; et al. DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging 2016, 8, 1844–1865. [Google Scholar] [CrossRef] [PubMed]
- Jylhävä, J.; Pedersen, N.L.; Hägg, S. Biological age predictors. EBioMedicine 2017, 21, 29–36. [Google Scholar] [CrossRef]
- Roetker, N.S.; Pankow, J.S.; Bressler, J.; Morrison, A.C.; Boerwinkle, E. Prospective Study of Epigenetic Age Acceleration and Incidence of Cardiovascular Disease Outcomes in the ARIC Study (Atherosclerosis Risk in Communities). Circ.-Genom. Precis. Med. 2018, 11, e001937. [Google Scholar] [CrossRef]
- Field, A.E.; Robertson, N.A.; Wang, T.; Havas, A.; Ideker, T.; Adams, P.D. DNA Methylation Clocks in Aging: Categories, Causes, and Consequences. Mol. Cell 2018, 71, 882–895. [Google Scholar] [CrossRef]
- Ryan, J.; Wrigglesworth, J.; Loong, J.; Fransquet, P.D.; Woods, R.L. A systematic review and meta-analysis of environmental, lifestyle, and health factors associated with DNA methylation age. J. Gerontol. Ser. A 2019, 75, 481–494. [Google Scholar] [CrossRef]
- Gems, D.; de Magalha, P.J. The hoverfly and the wasp: A critique of the hallmarks of aging as a paradigm. Ageing Res. Rev. 2021, 70. [Google Scholar] [CrossRef]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef]
- de Lima Camillo, L.P.; Asif, M.H.; Horvath, S.; Larschan, E.; Singh, R. Histone mark age of human tissues and cell types. Sci. Adv. 2025, 11, eadk9373. [Google Scholar] [CrossRef]
- Chang, E.; Harley, C. Telomere Length Furthermore, Replicative Aging In Human Vascular Tissues. Proc. Natl. Acad. Sci. USA 1995, 92, 11190–11194. [Google Scholar] [CrossRef]
- von Zglinicki, T.; Serra, V.; Lorenz, M.; Saretzki, G.; Lenzen-Grossimlighaus, R.; Gessner, R.; Risch, A.; Steinhagen-Thiessen, E. Short telomeres in patients with vascular dementia: An indicator of low antioxidative capacity and a possible risk factor? Lab. Investig. 2000, 80, 1739–1747. [Google Scholar] [CrossRef]
- von Zglinicki, T.; Martin-Ruiz, C. Telomeres as biomarkers for ageing and age-related diseases. Curr. Mol. Med. 2005, 5, 197–203. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Greider, C.W.; Szostak, J.W. Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 2006, 12, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Nordfjall, K.; Svenson, U.; Norrback, K.F.; Adolfsson, R.; Roos, G. Large-scale parent-child comparison confirms a strong paternal influence on telomere length. Eur. J. Hum. Genet. 2010, 18, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.T.; Seeboth, A.; Tsai, P.C.; Sun, D.; Quach, A.; Reiner, A.P.; Kooperberg, C.; Ferrucci, L.; Hou, L.; Baccarelli, A.A.; et al. DNA methylation-based estimator of telomere length. AGING-US 2019, 11, 5895–5923. [Google Scholar] [CrossRef]
- Oldham, K.B.; Spanier, J. The Fractional Calculus: Theory and Applications of Differentiation and Integration to Arbitrary Order; Academic Press: New York, NY, USA; London, UK, 1974. [Google Scholar]
- Miller, K.S.; Ross, B. An Introduction to the Fractional Calculus and Fractional Differential Equations; John Wiley and Sons Inc.: New York, NY, USA, 1993. [Google Scholar]
- Samko, S.G.; Kilbas, A.A.; Marichev, O.I. Fractional Integrals and Derivatives: Theory and Applications; Gordon and Breach Science Publishers: Yverdon, Switzerland; Philadelphia, PA, USA, 1993. [Google Scholar]
- Podlubny, I. Fractional Differential Equations. An Introduction to Fractional Derivatives, Fractional Differential Equations, Some Methods of Their Solution and Some of Their Applications; Academic Press: San Diego, CA, USA, 1999. [Google Scholar]
- Magin, R.L. Fractional Calculus in Bioengineering: Second Edition; Begell House Publishers: Redding, CT, USA, 2006. [Google Scholar]
- Mainardi, F. Fractional Calculus and Waves in Linear Viscoelasticity: An Introduction to Mathematical Models; Imperial College Press: London, UK, 2010. [Google Scholar] [CrossRef]
- Tarasov, V.E. Fractional Dynamics: Applications of Fractional Calculus to Dynamics of Particles, Fields and Media; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Magin, R.; Ortigueira, M.D.; Podlubny, I.; Trujillo, J. On the fractional signals and systems. Signal Process. 2011, 91, 350–371. [Google Scholar] [CrossRef]
- West, B.J. Fractional Calculus View of Complexity: Tomorrow’s Science; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2016. [Google Scholar]
- Mittag–Leffler, M.G. Sur la Nouvelle Fonction Eα(x). Comptes Rendus L’Académie Sci. 1903, 137, 554–558. [Google Scholar]
- Mittag–Leffler, M.G. Une generalisation de l’integrale de Laplace-Abel. Comptes Rendus L’Académie Sci. 1903, 137, 537–539. [Google Scholar]
- Mainardi, F.; Gorenflo, R. On Mittag–Leffler-type functions in fractional evolution processes. J. Comput. Appl. Math. 2000, 118, 283–299. [Google Scholar] [CrossRef]
- Kilbas, A.; Saigo, M.; Saxena, R. Generalized Mittag–Leffler function and generalized fractional calculus operators. Integral Transform. Spec. Funct. 2004, 15, 31–49. [Google Scholar] [CrossRef]
- Haubold, H.J.; Mathai, A.M.; Saxena, R.K. Mittag–Leffler functions and their applications. J. Appl. Math. 2011, 2011, 51. 298628. [Google Scholar] [CrossRef]
- Gorenflo, R.; Kilbas, A.A.; Mainardi, F.; Rogosin, S.V. Mittag–Leffler Functions, Related Topics and Applications; Springer Monographs in Mathematics; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef]
- Mainardi, F. On some properties of the Mittag–Leffler function Eα(-tα), completely monotone for t>0 with 0<α<1. Discret. Contin. Dyn. Syst.—Ser. B 2014, 19, 2267–2278. [Google Scholar] [CrossRef]
- Podlubny, I. Mittag–Leffler Function. MathWorks, Inc. MATLAB Central File Exchange, 2005. Available online: http://www.mathworks.com/matlabcentral/fileexchange/8738 (accessed on 1 July 2025).
- Garrappa, R. The Mittag–Leffler Function. MathWorks, Inc. MATLAB Central File Exchange, 2015. Available online: http://www.mathworks.com/matlabcentral/fileexchange/48154 (accessed on 1 July 2025).
- Snir, S.; Farrell, C.; Pellegrini, M. Human epigenetic ageing is logarithmic with time across the entire lifespan. Epigenetics 2019, 14, 912–926. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lapborisuth, K.; Farrell, C.; Pellegrini, M. Pseudotime analysis reveals exponential trends in DNA methylation aging with mortality associated timescales. Cells 2022, 11, 767. [Google Scholar] [CrossRef]
- McGovern, A.P.; Powell, B.E.; Chevassut, T.J. A dynamic multi-compartmental model of DNA methylation with demonstrable predictive value in hematological malignancies. J. Theor. Biol. 2012, 310, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Zagkos, L.; Mc Auley, M.; Roberts, J.; Kavallaris, N.I. Mathematical models of DNA methylation dynamics: Implications for health and ageing. J. Theor. Biol. 2019, 462, 184–193. [Google Scholar] [CrossRef]
- Nasrolahpour, H. Fractional Dynamics of DNA Methylation. ResearchGate 2024. [Google Scholar] [CrossRef]
- Dufek, G.; Katriel, G.; Snir, S.; Pellegrini, M. Exponential dynamics of DNA methylation with age. J. Theor. Biol. 2024, 279. [Google Scholar] [CrossRef]
- Nasrolahpour, H.; Pellegrini, M.; Skovranek, T. Fractional Calculus in Epigenetics. bioRxiv 2024. [Google Scholar] [CrossRef]
- Jaffe, A.E.; Gao, Y.; Deep-Soboslay, A.; Tao, R.; Hyde, T.M.; Weinberger, D.R.; Kleinman, J.E. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2016, 19, 40–50. [Google Scholar] [CrossRef] [PubMed]
| SAE | A | B | C | D |
|---|---|---|---|---|
| Exponential model | 15.8160 | 25.5782 | 15.6237 | 13.8314 |
| Mittag–Leffler model | 15.2298 | 23.8069 | 15.6238 | 12.9521 |
| SSE | A | B | C | D |
| Exponential model | 0.81503 | 1.73680 | 0.66498 | 0.56234 |
| Mittag–Leffler model | 0.77447 | 1.52290 | 0.66497 | 0.50813 |
| MAPE | A | B | C | D |
| Exponential model | 6.05810 | 6.52110 | 3.63370 | 9.70770 |
| Mittag–Leffler model | 5.38320 | 5.82850 | 3.63390 | 9.39840 |
| A | B | C | D | |
| Exponential model | 0.93612 | 0.64137 | 0.55855 | 0.75920 |
| Mittag–Leffler model | 0.93930 | 0.68555 | 0.55856 | 0.78241 |
| A | B | C | D | |
| Exponential model | 0.93601 | 0.64078 | 0.55782 | 0.75880 |
| Mittag–Leffler model | 0.93910 | 0.68451 | 0.55710 | 0.78170 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasrolahpour, H.; Pellegrini, M.; Skovranek, T. Fractional Calculus in Epigenetics: Modelling DNA Methylation Dynamics Using Mittag–Leffler Function. Fractal Fract. 2025, 9, 616. https://doi.org/10.3390/fractalfract9090616
Nasrolahpour H, Pellegrini M, Skovranek T. Fractional Calculus in Epigenetics: Modelling DNA Methylation Dynamics Using Mittag–Leffler Function. Fractal and Fractional. 2025; 9(9):616. https://doi.org/10.3390/fractalfract9090616
Chicago/Turabian StyleNasrolahpour, Hosein, Matteo Pellegrini, and Tomas Skovranek. 2025. "Fractional Calculus in Epigenetics: Modelling DNA Methylation Dynamics Using Mittag–Leffler Function" Fractal and Fractional 9, no. 9: 616. https://doi.org/10.3390/fractalfract9090616
APA StyleNasrolahpour, H., Pellegrini, M., & Skovranek, T. (2025). Fractional Calculus in Epigenetics: Modelling DNA Methylation Dynamics Using Mittag–Leffler Function. Fractal and Fractional, 9(9), 616. https://doi.org/10.3390/fractalfract9090616










