1. Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder that significantly impacts an individual’s motor abilities and daily functioning. It is characterized by a variety of symptoms, including tremor, muscle rigidity, bradykinesia, and impaired balance and coordination. According to the World Health Organization (WHO) [
1], PD affects approximately 1–2 per 1000 people currently, with its incidence increasing with age. Globally, statistics indicate that the incidence of disability and mortality associated with PD exceeds that of any other neurological pathology. Since 2000, the prevalence of PD has doubled, and WHO estimates report an increase of 81% and more than 100% in disability-adjusted deaths and deaths caused by this neurological disorder. Furthermore, the stigma of the disease and the psychological and logistical burdens placed on the families of the patients highlight the urgent need for improved healthcare and social support systems, including more efficient diagnostic processes.
Recent studies have shed light on various aspects of Parkinson’s disease, revealing intriguing insights into its complex nature. For example, emerging research suggests a potential link between gut health and PD. The gut-brain axis, a bidirectional communication system between the gastrointestinal tract and the brain, has gained attention in PD research [
2]. Researchers propose that changes in the gut microbiota could impact the onset and progression of PD, suggesting that this axis could offer novel insights and therapeutic targets for the disease.
Furthermore, the role of neuroinflammation in PD has attracted increased attention. Chronic brain inflammation, primarily driven by cells such as microglia and astrocytes, plays a crucial role in PD pathology [
3]. This inflammation, often initiated by misfolded alpha-synuclein proteins, contributes significantly to the degeneration of neurons critical for motor control. Developing therapeutic strategies that target this inflammation could potentially slow or stop the progression of PD.
The exact causes of Parkinson’s disease are not yet fully understood, but are believed to stem from a combination of genetic and environmental factors [
4]. Researchers have made significant progress in understanding the underlying mechanisms and pathophysiology of the disease, leading to the development of various diagnostic and treatment approaches [
5,
6]. Non-pharmacological interventions have also been systematically reviewed in the literature, highlighting the relevance of tailored therapeutic strategies in PD management [
7]. Despite these advances, the diagnosis of PD still relies heavily on clinical evaluations considering symptoms, medical history, and physical examination findings [
8]. The absence of a definitive diagnostic test complicates early detection and timely intervention of the disease [
9].
The challenge of early diagnosis has sparked innovative research in the field of medical technology. More efficient and accurate diagnostic methods are needed to facilitate early identification of the disease [
10]. Currently, advances in therapeutic interventions and research into new technologies aim to revolutionize early diagnosis and treatment strategies [
11]. In particular, Artificial Intelligence (AI), and particularly Machine Learning models, have emerged as a promising field, offering substantial support in diagnosis and screening applications.
Artificial intelligence’s role in healthcare has significantly expanded, with Machine Learning, especially Deep Learning models, playing a pivotal role in analyzing complex datasets. These models have been effectively applied in various domains, including medical imaging [
12,
13], audio analysis [
14], and biomedical signal processing [
15,
16]. The use of DL models can enhance diagnostic precision and operational efficiency, empowering healthcare professionals to make more informed decisions.
Gated Recurrent Neural Networks (RNNs), which encompass architectures like Long-Short Term Memory (LSTM) and Gated Recurrent Unit (GRU), have demonstrated potential across a range of medical applications. Their effectiveness as classifiers for medical time series data [
17,
18] indicates that they could be particularly valuable in the early diagnosis of Parkinson’s disease, where timely and accurate detection is crucial.
This research study aims to analyze the effectiveness of AI models based on Gated RNNs as diagnostic support tools for early diagnosis of Parkinson’s disease utilizing plantar insole pressure sensors. By analyzing subtle alterations in gait and plantar pressure patterns, this system aims to accurately identify Parkinson’s disease in its early stages. Such an approach could greatly improve early detection and intervention efforts, potentially leading to significant improvements in patient outcomes and quality of life.
Advancements in wearable technology have revolutionized the monitoring and management of Parkinson’s disease. Equipped with sensors to track movement patterns and vital signs, wearable devices facilitate continuous health monitoring beyond the confines of clinical settings. This technology not only aids in the early detection of motor symptoms, but also facilitates the development of customized treatment plans [
19]. Integrating these devices with AI algorithms could lead to real-time patient-specific management of Parkinson’s disease, enhancing treatment efficacy.
The remainder of the manuscript is organized as follows.
Section 2, Related Works, reviews the existing literature pertinent to the use of Gated Recurrent Neural Networks in medical applications, particularly for Parkinson’s disease.
Section 3 describes the materials and methods used in this study, including the dataset for training and evaluation, the specifics of the two types of RNN architectures employed, the architectures considered, and the criteria for evaluating their effectiveness.
Section 4 presents the results and discussion of the Deep Learning models that were trained. Finally,
Section 5 concludes the paper with a summary of the findings and discusses potential avenues for future research.
4. Results and Discussion
The hyperparameter optimization results indicate that a high number of nodes leads to overfitting. Furthermore, the learning rate values that yielded the best results were found to be between 0.005 and 0.001. Regarding batch size, high values were observed to cause lower performance in the validation subset, once again being interpreted as favoring overfitting. Regarding dropout, the best results were obtained with values between 0.1 and 0.3. The best results obtained in the first phase are shown in
Table 3.
LSTM networks show slower convergence than GRU networks. Some two-layer LSTM models achieved high performance, but convergence was slower. However, the best results were obtained from training models with a single-layer GRU architecture. During training, it was observed that models with a single recurrent layer generally suffer less from overfitting due to the lower complexity of the architecture. However, models with a single LSTM layer reached lower performance values. Despite some two-layer LSTM architectures exhibiting similar performance, due to their slower convergence, increased tendency to overfit, and higher complexity, the analysis of a two-layer LSTM architecture was not considered.
The final architecture chosen had one GRU layer with 8 nodes. The dropout was 0.2 and the batch size was 32 samples used to calculate the training error. The learning rate used was 0.001. We selected this model after analyzing all the mentioned models for their convergence and prediction accuracy using the validation subset.
The results obtained with this model using the test subset are summarized in
Table 4. The first row of the table indicates the metrics obtained when classifying each sample from the test subset, which is consistent with the results of the “one GRU layer” in
Table 3 (the selected candidate).
However, to delve deeper into the potential of the model as a diagnostic support tool, the performance of the model was analyzed by classifying subjects with Parkinson’s disease, rather than individual gait samples. To achieve this, the model was used to classify all the samples in the test set from each subject such that a subject was classified as having Parkinson’s disease if at least half of their samples were identified as Parkinson’s-positive. This simple majority criterion was chosen because it is the most widely used voting strategy in similar biomedical-signal studies and because it preserves symmetry between the two classes when no disease-prevalence priors are available. So, the results are shown in the same table from the point of view of the subject classification (
Table 3, second row). Because the same number of samples are not available for each subject, the results of these two rows do not align. This discrepancy arises because the number of samples is not uniform across subjects. Indeed, subjects for whom the aggregate classification improved tended to contribute a higher number of samples, highlighting the benefit of the aggregation rule.
To further assess the robustness of this aggregation rule and its implications for diagnostic performance, we carried out a post-hoc sensitivity analysis to examine how different voting thresholds affect diagnostic performance. When the cut-off is lowered to forty percent, one additional control subject is falsely labelled as PD, whereas raising the threshold to sixty percent causes exactly one PD subject to switch to the control class. Accuracy changes by less than two percentage points across the 40–60% range, and recall remains above eighty-five percent at every threshold. These results indicate that the model is not unduly sensitive to the particular fifty-percent boundary. From a clinical perspective, this majority-vote rule reflects a realistic usage scenario: in an at-home setting, a wearable device would record hundreds of gait segments per day, allowing clinicians to choose a decision threshold that balances false negatives and false positives according to individual risk profiles. By documenting the threshold’s stability and by making the parameter explicit in the manuscript, we provide a transparent basis for such future fine-tuning.
It can be observed that following this criterion leads to higher performance. Based on these results, we interpret that although the model may misclassify more individual gait segments, its performance improves when considering the entire gait record.
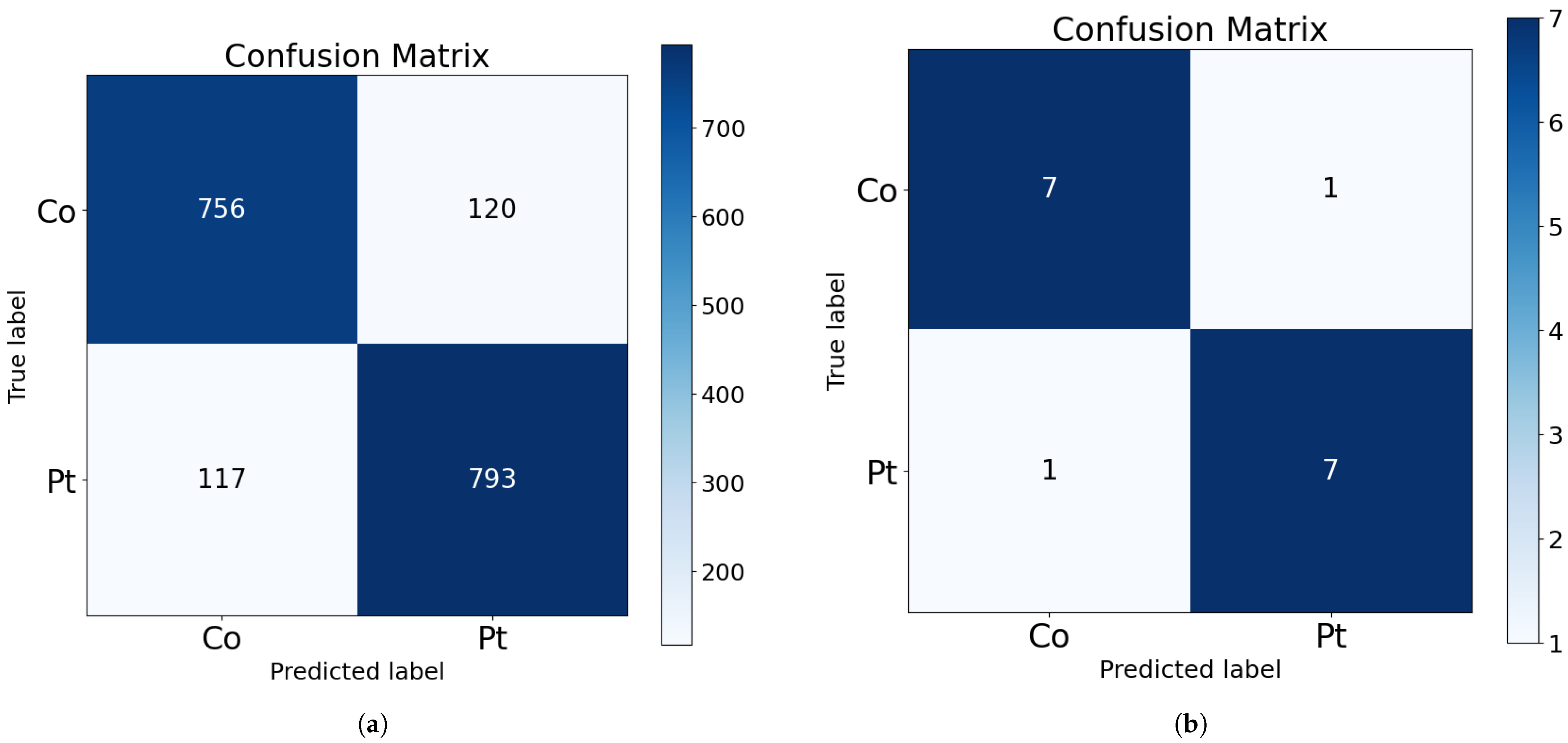
Their corresponding confusion matrices are shown in
Figure 2. As shown in the bottom part of the figure, only one subject from each class is misclassified in the test subset. This corresponds to approximately 100 misclassified samples from each class (as visible in the upper part of the figure).
The confusion matrix illustrated in
Figure 2b demonstrates an improved true positive rate and a more acceptable balance of false positives, in percentage terms, when compared to the results of the classification of individual samples. This outcome is more favorable for the use of the model as a diagnostic and screening tool.
Finally,
Table 5 presents the results obtained with the selected architecture using K-fold cross-validation with K = 6. For the case of sample classification (upper part of the table), the results obtained on the training set were very high for all the considered dataset combinations. The accuracy results exceeded 93% for all training subsets. Furthermore, when models were trained focusing on the recall metric, values above 98% were achieved for all training subsets, which is promising, suggesting the classifier’s potential as a screening tool.
For the results of each fold on the test subset, a decrease in performance values was observed, similar to those seen in the initial phase of this study. This suggests a dependency on the specific data used for training and a tendency to overfit to specific training characteristics. However, there is a significant variation in performance across specific folds, such as the first fold, suggesting that certain subjects may not exhibit the common gait characteristics prevalent in the rest of the dataset.
As previously done, performance metrics were extracted by classifying subjects based on all samples available for each individual. The results are shown in
Table 5 (bottom part). With few exceptions, the results obtained demonstrated improvements compared to the classification of individual samples.
The best result was obtained with fold 3, achieving more than 90% accuracy for the classification of samples and more than 93% precision for the classification of subjects. The average results obtained for the final test of the best candidate were approximately 85% accuracy and nearly 87% recall for the test subset when working with samples. For the case of subject classification, the average results were over 86% accuracy and nearly 100% recall for the test subset.
Regarding the worst results obtained after cross-validation tests, these were observed with fold 1. For the case of sample classification, an accuracy of 78% and a recall of 77% were obtained for the test subset. And, for the case of classification of subjects, an accuracy and a recall of 81% were obtained. Thus, this implies a standard deviation of approximately 5% for samples and subjects, which is a relatively low value and acceptable for a diagnostic aid system.
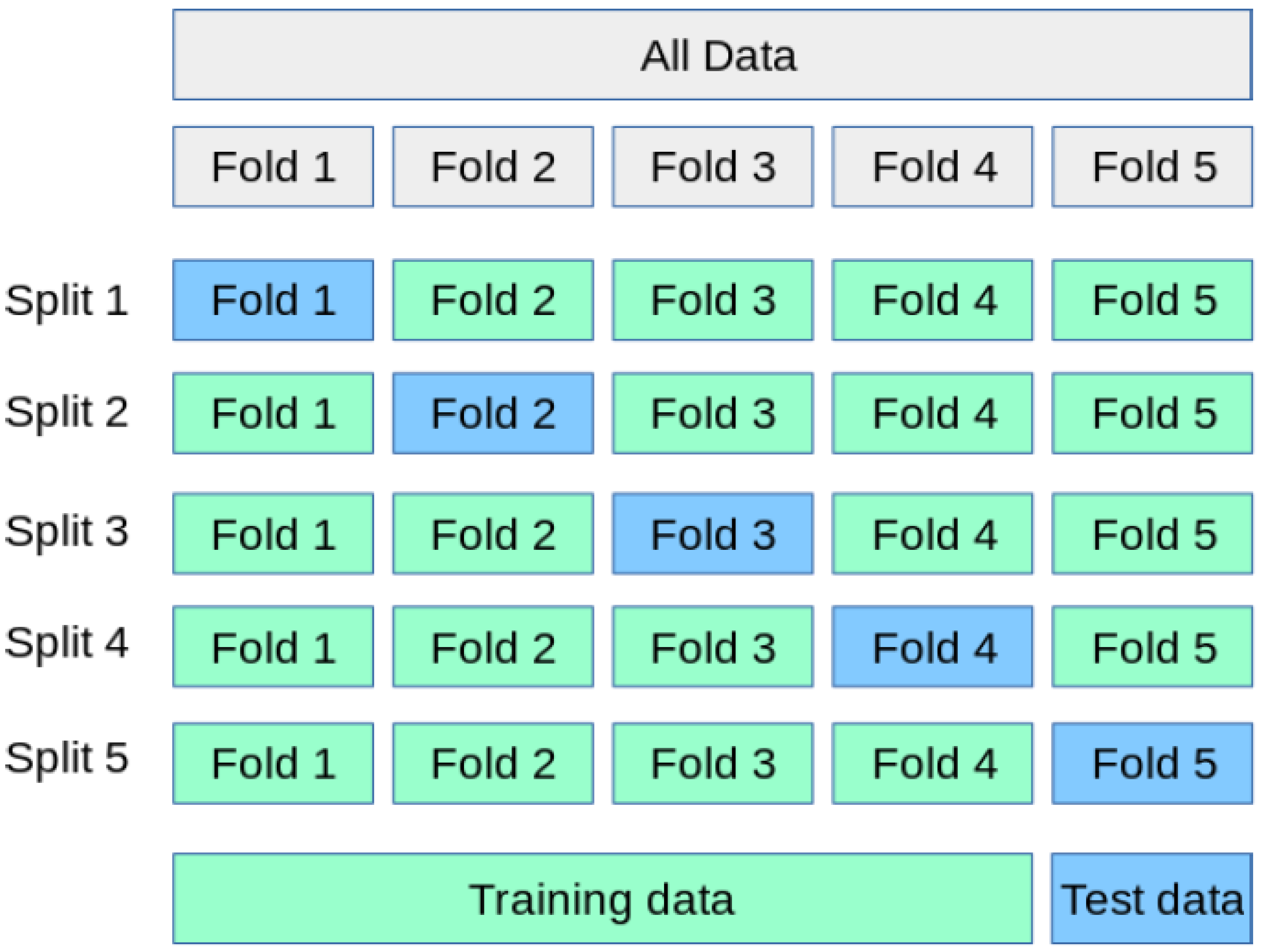
It is important to highlight that the variability observed in the K-fold cross-validation results can be attributed to the data partitioning strategy. To prevent data leakage, K-fold partitions were created at the subject level rather than the sample level. This implies that each validation fold could contain a set of subjects with gait patterns significantly different from those in the training set, which, in turn, introduces fluctuations in performance metrics across different folds. Despite these variations, the classifier’s accuracy never dropped below 78%, underscoring the overall robustness of the approach. Nevertheless, we acknowledge that to ensure greater stability and generalizability in real-world deployment scenarios, the use of larger and more balanced datasets will be required in future work.
In general, the results indicate that patients with Parkinson’s symptoms exhibit a very characteristic walking pattern. While this pattern may not be easily detectable in all patients, a system based on artificial intelligence techniques has proven useful in these circumstances, as, when pooling the correctly classified subjects from the training, validation, and test subsets, the combined accuracy is well over 95%.
Finally, to compare the results of this work with those obtained in previous studies, a bibliographic search was conducted for journal articles published in the last 10 years that utilized the same dataset as this study. The search identified a total of three articles with generally favorable results, and a summary of all of them is summarized in
Table 6.
In this table, to facilitate comparison, the average and the best results obtained in this work were included as the last two rows.
Analysis of the results from previous studies indicates that high accuracy results were achieved in all of them. This means that the dataset is highly representative and that the patterns of Parkinson’s patients are readily detectable.
However, in these previous studies, different techniques were employed to classify and validate the results, which underscores the unique value of the present work.
Given the similar accuracy in many of the works, execution time, a common performance metric for hardware systems, could be used for comparison, although none of the identified papers present such a study. Therefore, a thorough comparison would ideally involve implementing the algorithms and classifiers from the previous papers on the same device to measure, under identical hardware constraints, the execution times of each of the models.
However, previous works often do not (in most cases) provide a detailed description of their classifiers (only a theoretical and/or graphical description), making their practical implementation challenging.
For this purpose, we refer to a previous study conducted by some of the current authors, which provides a comparison of execution times between classifiers based on classical neural networks and recurrent neural networks [
31]. These experiments were performed under the same hardware constraints (processor, memory, gpu), the same software constraints (programming language, libraries) and the same data constraints (dataset, subset split, and preprocessing).
In this work, average running (classification) times were presented for artificial neural networks with 1 and 2 hidden layers, as well as for recurrent neural networks of LSTM and GRU types (with one and two layers each). The results of the execution times are summarized in
Table 7.
From
Table 7, it is observed that a classification algorithm with one GRU layer takes approximately 10 times longer than a classical ANN with one hidden layer (a factor also applicable to a single LSTM layer), whereas two LSTM layers take 15 times longer. Thus, assuming a worst-case scenario for this study, we hypothesize that a fully connected layer has an execution time of
T and, based on
Table 7, one GRU layer has an execution time of
, and two LSTM layers have an execution time of
. While more complex processing layers (such as convolutional layers and pooling layers), their execution time will be longer than
T, but we will consider the most conservative case for our analysis and assume that all non-recurrent layers will have an execution time of
T.
Taking these assumptions into account,
Table 8 summarizes the hypothetical execution times for each work under the same conditions.
From the first article [
20], the comparison is complex, as the number of dense layers included in its ANN is not detailed in the manuscript. Furthermore, it is not possible to estimate the complexity of the preceding LBP feature extraction layer. All that can be seen in the paper is the presence of 4 processing blocks (one of which is the ANN network of unknown complexity). Even so, this work presents several unspecified unknowns: since it does not use cross-validation, it is unclear how many training runs were conducted to find a division between training, validation, and test that provides the shown results; additionally, the division percentages of the subsets are not indicated (nor whether a validation set was used); furthermore, it is not specified whether the accuracy results presented are from the test set or the total dataset (which would inflate the actual classification results of the system); and finally, the best accuracy results obtained do not surpass the best results achieved in our work.
Continuing with the work by [
21], in addition to the issue of not using cross-validation, this work lacks reported accuracy results (only recall is provided). Considering this metric, the results are similar to ours. However, comparing the complexity of the classifier system as indicated in
Table 8, the optimistic estimate indicates a complexity of 200% compared to our classifier.
Regarding the work of [
22], the accuracy and recall results are notably high; however, only the best outcomes obtained with a
K = 10-fold cross-validation are illustrated, and no information is provided on the distribution of the data, which limits comparative analysis. In terms of computational complexity, again due to the nature of the implementation, comparison becomes complex. Random Forest can be implemented with high efficiency, even on embedded systems. However, for improved precision, the proposed model adjusts the information obtained from each step based on certain neighbors within the dataset. This requires the dataset to be accessible during execution, which may be unfeasible for embedded systems due to resource limitations. Furthermore, feature extraction involves computationally expensive calculations.
The work of [
23] presents results that surpass ours regarding accuracy and recall. Similarly, the validation mechanism, utilizing cross-validation, is highly similar to ours. On the other hand, the best model presented in [
32] also achieves superior precision, in this case without employing cross-validation. However, if we compare these works in terms of execution time, the optimistic forecast made in
Table 8 indicates that they exhibit execution times approaching 600% and 200%, respectively, relative to our work.
These results show that our classifier achieves nearly the best classification results while maintaining a leading position in computational efficiency, as for comparable results, we achieve a substantial improvement in execution time. This aspect is not critical when utilizing high-capacity computing equipment (such as a hospital computer), but it is crucial for integrating the classifier into a wearable device.
To further support the feasibility of deploying our model in wearable devices, we conducted a simulation using the X-Cube-AI framework provided by STMicroelectronics. The analysis was configured for an STM32F411RE microcontroller (Cortex-M4), and the results confirmed that the proposed GRU-based classifier fits within the memory and computational constraints of the device. Specifically, the model size was estimated at approximately 196 KB, which fits comfortably within the available flash memory. The estimated inference time was around 120 milliseconds, a plausible response time for continuous monitoring scenarios, where new samples are acquired during each inference cycle. While a direct comparison to all state-of-the-art models on this specific hardware was infeasible due to various constraints, including the impracticality of re-implementing every architecture and limitations of the deployment toolchain, these results are consistent with those reported in a previous study [
17], in which a similar GRU-based architecture was successfully deployed on a real STM32 embedded system. The reported memory footprint and inference speed suggest that our solution significantly reduces the computational overhead compared to larger, multi-layer architectures or those reliant on complex feature engineering, making it well-suited for resource-constrained wearable applications. These findings reinforce the potential of our approach for real-time execution on low-power platforms.
Furthermore, based on a comparative assessment of model complexity, we estimate that alternative architectures proposed in the literature—particularly those with 2× to 6× higher computational demands—would likely require more capable embedded systems with increased memory and processing speed to achieve real-time inference. While such systems may deliver slightly higher classification accuracy, they would also entail increased energy consumption and hardware cost. These trade-offs are critical when selecting an appropriate model for deployment in wearable diagnostic applications, where efficiency and portability are essential. This aspect has previously been addressed in previous work by this research group, such as [
33], demonstrating that low computational cost classification algorithms can be run in real time on embedded systems with low computational resources.
Limitations and Future Work
Despite the promising results presented in this study, several limitations should be acknowledged. Firstly, we did not carry out a systematic analysis of misclassified instances; therefore, the specific gait patterns leading to false positives and false negatives remain unclear. Analyzing these misclassifications could reveal valuable insights into model weaknesses and guide improvements. Secondly, our evaluation relied exclusively on the “Gait in Parkinson’s Disease” dataset. Although this dataset is comprehensive in terms of sensor coverage, its subject pool is limited and unevenly distributed between disease severity levels: certain stages are represented by only one participant. Because our K-fold splits are performed at the subject level to avoid data leakage, folds that isolate these unique participants in the test set can exhibit lower accuracy, reflecting the model’s difficulty in generalising to under-represented gait patterns. Future studies should incorporate larger and better balanced cohorts—ideally with explicit metadata on Parkinson’s severity—to mitigate this issue and improve generalizability.
Furthermore, while the dataset contains rich clinical metadata such as Hoehn & Yahr stage, medication status, and comorbidities, these variables were not utilized in the current study. This was a deliberate choice to align with our envisioned wearable implementation, which relies solely on plantar-pressure signals in everyday use. Consequently, omitting these variables limits our ability to assess how model performance may vary with disease severity or medication state. However, we acknowledge that leveraging such information holds significant potential for future extensions aiming to improve accuracy or enable more personalized models.
Moreover, further research could explore the integration of multimodal sensor input beyond plantar pressure sensors, potentially enhancing classification accuracy and robustness.
The present study primarily focuses on demonstrating the technical feasibility that plantar-pressure data alone can support reliable classification of Parkinson’s gait patterns; however, it is not intended to replace or replicate comprehensive neurologist evaluation. In practice, a wearable implementation would primarily operate as an early-screening or continuous-monitoring aid. When the algorithm detects a sustained pattern suggestive of Parkinson’s disease, the device would notify both the user and a healthcare professional, prompting a formal clinical examination (e.g., UPDRS scoring) rather than delivering a stand-alone diagnosis. Such integration could accelerate referral for specialist assessment and facilitate objective, longitudinal tracking of gait changes between clinic visits, thereby complementing neurologist judgement rather than competing with it.
It is also crucial to acknowledge the practical consequences of misclassifications. While our best model achieves solid subject-level performance (accuracy ≈ 0.88, precision ≈ 0.94, recall ≈ 0.88; see
Table 4), any misclassification could have significant implications. False negatives are of particular concern because an undetected patient may miss out on timely intervention, which could lead to worsening disease progression. False positives, while medically less dangerous, can still impose psychological stress and lead to unnecessary follow-up testing. Therefore, this classifier is intended strictly as a screening aid and must always be followed by a comprehensive clinical evaluation. In any real-world deployment, results would be reviewed by a qualified clinician before any diagnosis is communicated, ensuring a human-in-the-loop safeguard and adherence to ethical guidelines for diagnostic support systems. Prospective validation under appropriate ethical approval will be an essential next step.
Finally, although we conducted detailed simulations confirming the feasibility of deployment on embedded hardware, we did not perform a full hardware implementation on an actual wearable device. Such an implementation and validation in real-world conditions represents an essential next step. It would allow us to observe the behavior of the models when integrated and executed over extended periods on embedded platforms designed for wearable applications. This would help confirm whether the performance metrics observed in this study remain stable in practice and would also enable the measurement of actual power consumption and device autonomy, which are critical parameters for real-world usability. A full treatment of other real-world engineering aspects, such as comprehensive power consumption analysis, sensor drift mitigation, on-device inference latency optimization, and robust handling of motion artifacts, lies beyond the scope of this initial study, but will be thoroughly addressed during the forthcoming hardware-integration and real-world validation phases.