1. Introduction
The COVID-19 pandemic has presented unprecedented challenges to healthcare systems worldwide, necessitating rapid and accurate methods for diagnosis and severity assessment. Since late 2019, COVID-19 has caused more than 7 million deaths and infected more than 704.7 million people globally as of Aug, 2024 [
1]. Although RT-PCR remains the primary method for identifying current infections, imaging techniques such as chest X-rays have proven valuable for both diagnosis and severity classification [
2,
3,
4].
Chest X-rays are preferred over CT scans for COVID-19 assessment due to their lower cost, reduced risk of cross-infection, and wider availability [
3,
5]. However, the visual interpretation of chest X-rays is subject to inter-observer variability and can be time-consuming, especially during periods of high patient influx. This has led to increased interest in developing automated methods for COVID-19 severity classification using chest X-rays.
The bias towards severe cases in both COVID-19 severity classification and binary diagnosis models developed to date poses a challenge to accurately assess the full spectrum of COVID-19 cases. The high accuracy of binary COVID-19 diagnostic models is due to the abundance of training examples of healthy individuals without pneumonia indications, as well as images from critical care settings [
6,
7,
8]. In the case of COVID-19 severity classification, the model’s classification accuracy in classifying the extreme cases was greater than in classifying the mild cases [
9], which shows that the model was biased toward severe conditions. The true medical value of a machine model lies in its capacity to detect situations that fall somewhere between the extremes. In addition, identifying the severity level can be useful in monitoring the progression of the disease.
Grading the severity of COVID-19 positive cases can assist in reducing the problem that arises due to the unusable clinical value of the classification models [
9]. Most of the classification models in this area focus on identifying whether a patient is COVID-19 positive or not. Binary classification is essential as it could help us do the initial screening and quarantining. However, this use could be overshadowed if the model is fitted on a dataset that is highly discriminated between negative cases and infected severe cases. That might be the case with several studies [
6,
7,
8,
10,
11] on chest X-ray binary classification, as they have claimed an accuracy greater than 90%.
Severity classification methods have utilized convolutional neural networks (CNNs) such as ResNet50 and DenseNet-based approaches for computer vision tasks. Although CNNs have dominated the field since the introduction of AlexNet [
12], Vision Transformers (ViT [
13]) have recently shown satisfactory performance on large datasets. However, ViT has an issue with translation equivariance and requires a large dataset to learn object translation [
14]. In contrast, CNNs lack the global receptive field and adaptive input weighting properties of self-attention found in ViT. These properties allow ViT to see the input at a single glance and understand complex correlations between input data [
14].
Persistent homology excels in capturing robust and scale-invariant global topological characteristics of medical images, such as connected components and holes. Unlike CNNs and Transformers, PH can reliably encode structural relationships that are less sensitive to variations in orientation, illumination, or minor spatial perturbations. This provides stable descriptors that are particularly valuable in medical imaging tasks where anatomical structures or pathological abnormalities manifest distinct topological patterns [
15,
16].
CNNs inherently capture critical local features and textures in medical images, enabling effective extraction of spatial patterns crucial for disease classification. Their inherent translation equivariance ensures that local anatomical variations or lesion locations across images do not degrade the classification performance significantly, providing stable and generalized representations critical in clinical diagnosis [
17,
18].
Vision Transformers provide the essential ability to model global dependencies and adaptive attention across the entire image. This global contextualization capability allows for the accurate modeling of clinical features dispersed throughout an image, overcoming CNN limitations related to restricted receptive fields. Thus, Transformers significantly enhance the model’s sensitivity to subtle but clinically important relationships across distant regions within chest X-rays or other medical imaging modalities [
13,
19].
This hybrid approach is justified over single or simpler combinations.
Persistent homology alone struggles with instability issues related to minor data variations, requiring stabilization or complementary features for reliable clinical applications [
16].
CNN-only models lack a global receptive field and might overlook globally dispersed pathological features, limiting clinical utility where comprehensive context is crucial [
18].
Transformer-only architectures, despite their global contextual understanding, require substantial data and may struggle to accurately capture nuanced local textures essential for detailed diagnosis without CNN-like inductive biases [
19].
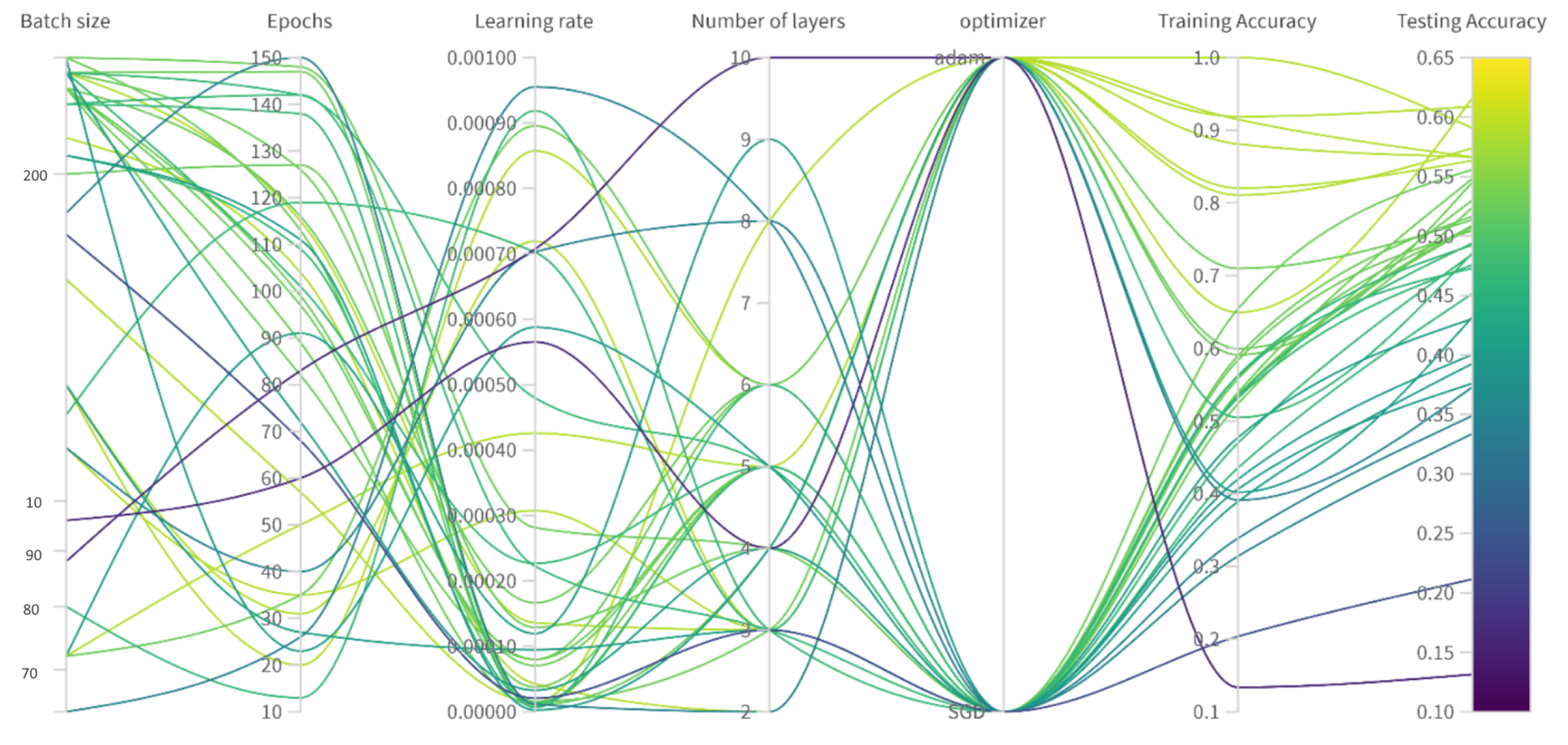
This study aims to identify indicators of COVID-19 by locating opacity changes and abnormalities by integrating these three methods. The proposed hybrid approach leverages their respective strengths to achieve robust, globally aware, and locally sensitive feature extraction suitable for highly reliable and clinically meaningful diagnosis, clearly outperforming simpler or individual methodologies. Two research questions were undertaken: one to ask how topological feature extraction techniques improve the performance of the COVID-19 severity classification model, and the second is to see how the proposed model performs compared with ResNet-50, VGG16, and SVM.
The main contributions of this study are as follows:
Development of a hybrid feature extraction technique that integrates topological data analysis with state-of-the-art deep learning methods.
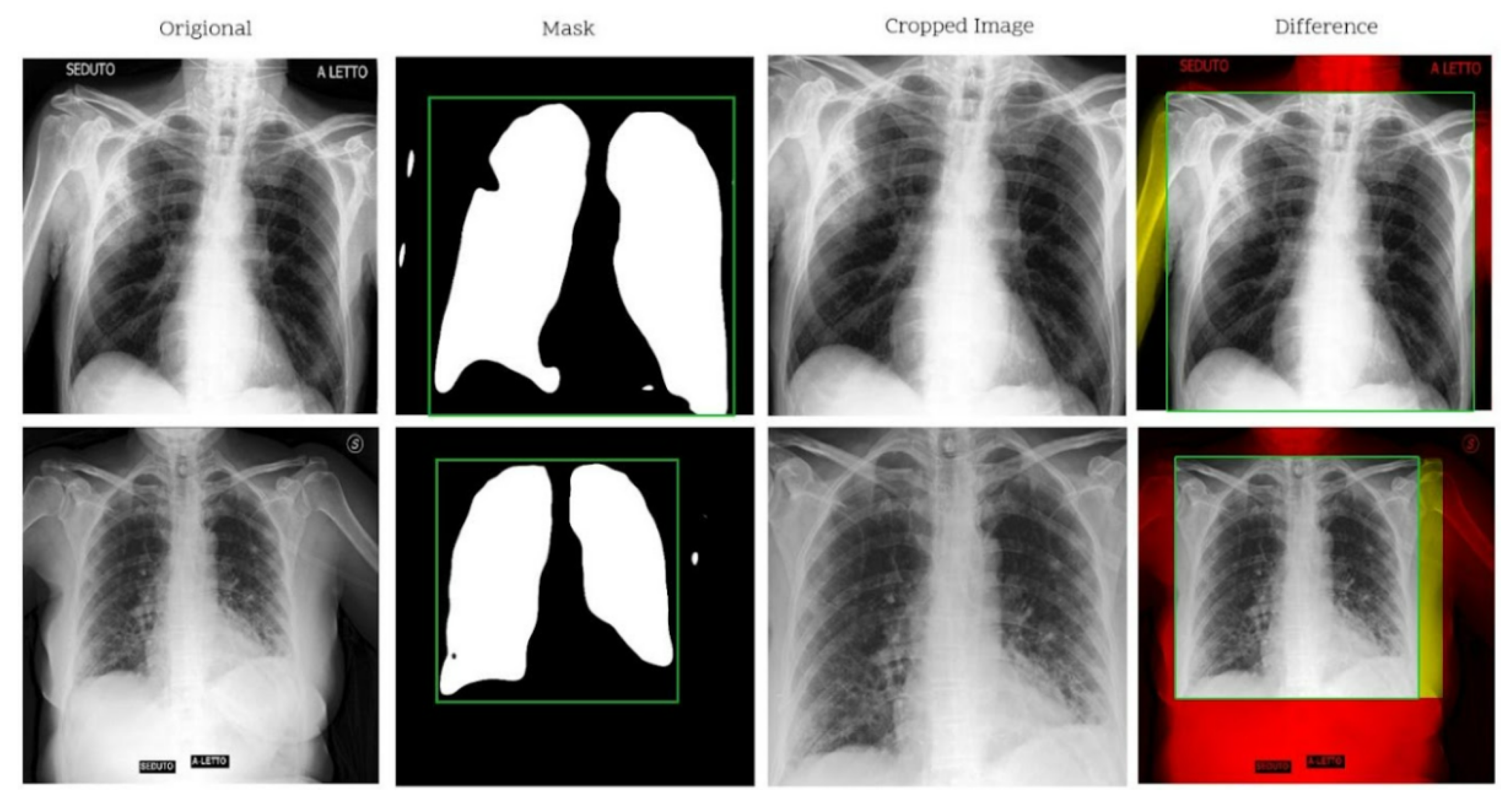
Introduction of a modified segmentation-based cropping technique that improves lung region isolation.
Comprehensive comparison of the proposed method with existing approaches, demonstrating improved accuracy and reduced bias across severity levels.
Analysis of the contribution of persistent homology features to the overall classification performance.
The rest of this paper is organized as follows:
Section 2 reviews related work in COVID-19 severity classification.
Section 3 describes the proposed methodology in detail.
Section 4 presents the experimental setup and results.
Section 5 discusses the findings and their implications. Finally,
Section 6 concludes the paper and suggests directions for future research.
2. Related Works
Most of the works carried out involve the detection of COVID-19 by using datasets containing a few hundred images and/or by transfer learning existing models to differentially diagnose COVID-19 from other types of pneumonia. The clinical utility of a chest X-ray is clearer in disease progression monitoring and severity assessment than in differential diagnosis [
20].
In [
21], a private dataset is produced by labeling nine publicly available datasets using two radiologists with 10 years of experience. Four classes that are mild, moderate, critical, and severe with 1000, 950, 600, 710 chest X-ray images were scored using a modified RALE score having opacity score, and lung involvement score. An end-to-end CNN classifier with 16 layers was trained after the hyperparameters were determined using grid search optimization. The authors compared the trained model with VGG16, AlexNet, ResNet-34, and ResNet-101 and found out that it outperformed all of them with 95% accuracy.
Although the authors of [
21] claimed to have adopted the RALE score, they have failed to clearly state the ranges in which the classes are to fit. Furthermore, despite the fact that the dataset was built from publicly available data, it has not yet been released to the public. The training has followed validation phases where they verified the model’s performance on unseen data. However, the dataset could have been balanced and segmented to have the lung region only to prevent overfitting.
In [
9], the need for a new dataset where the images come from the same machine having similar acquisition parameters was justified. Using the RALE scoring technique, 426 chest X-ray images were labeled as Normal, Mild, Moderate, and Severe by four radiologists. The authors applied the U-net segmentation model to crop out unwanted regions of the lung. Once the lung region was masked out, the maximum (and minimum) values of the lung plus 2.5% of the length/width were used to calculate the cropping rectangle. Synthetic augmentation (using class-inherent transformation network) was applied to expand the dataset. ResNet50 classifier was used to score an accuracy of 76.18%.
In [
22], 65 chest X-ray images from 48 patients were labeled by two radiologists with five and seven years of experience. Alveolar opacity patterns, which are circle-shaped opacities, and interstitial opacity patterns, which are linear opacities extending from the hilum to the lung parenchyma, are scored independently from 0 to 3 depending on the size and region of the involvement. A modified U-Net segmentation model (with EfficientNet as a base model) trained on 1048 publicly available images was used to specify the region of interest. Densenet121 pretrained on the ImageNet dataset was fine-tuned to classify the newly constructed dataset. The authors reported the models’ accuracy to be 78.5% in classifying alveolar opacities and 90.7% in classifying interstitial opacities.
In [
20], a much larger dataset of 5000 Chest X-ray images was used to predict the Brixia score. The authors used a modified U-Net to segment the lung. This was followed by an alignment procedure that rotates, centers, and crops the lung into an upright lung position. The score prediction was performed by feeding the aligned image into a sequence of convolution, feature pyramid network, and finally, global average pooling, followed by SoftMax activation layers. It was able to predict the scores with a correlation coefficient of 0.86.
While significant progress has been made in automated COVID-19 chest X-ray analysis, addressing dataset bias, improving generalization, and standardizing severity assessment methods remain critical challenges. Future work should focus on developing more robust, clinically relevant models that can reliably assist in patient care across diverse healthcare settings.
5. Discussion
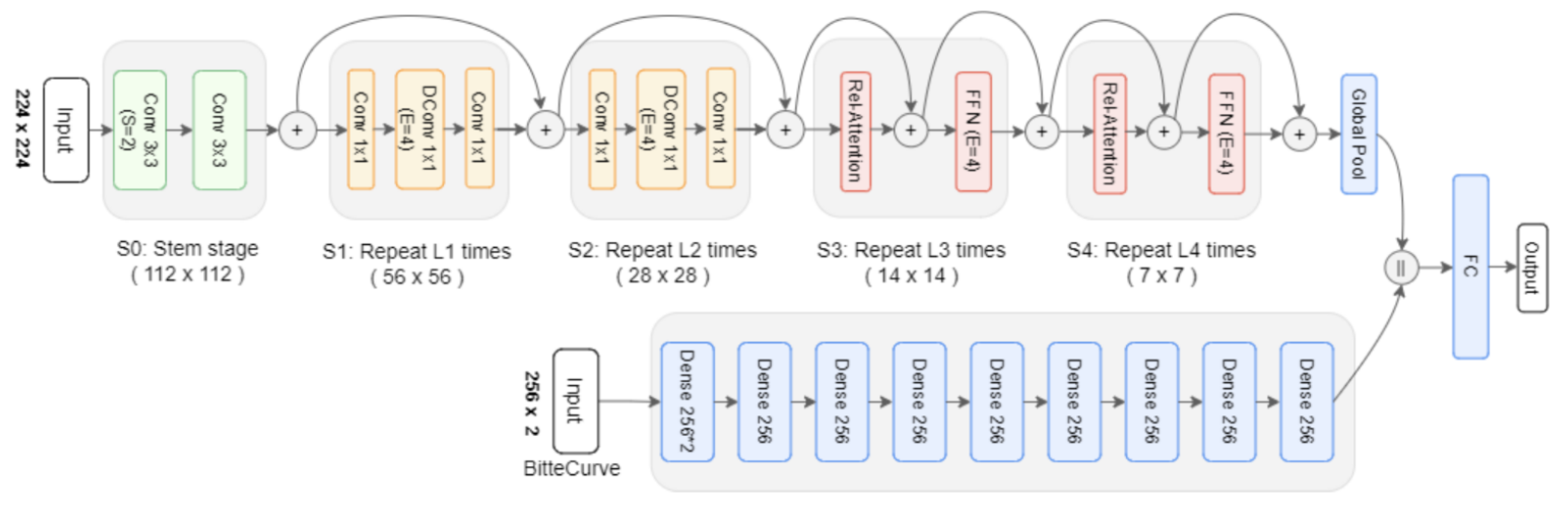
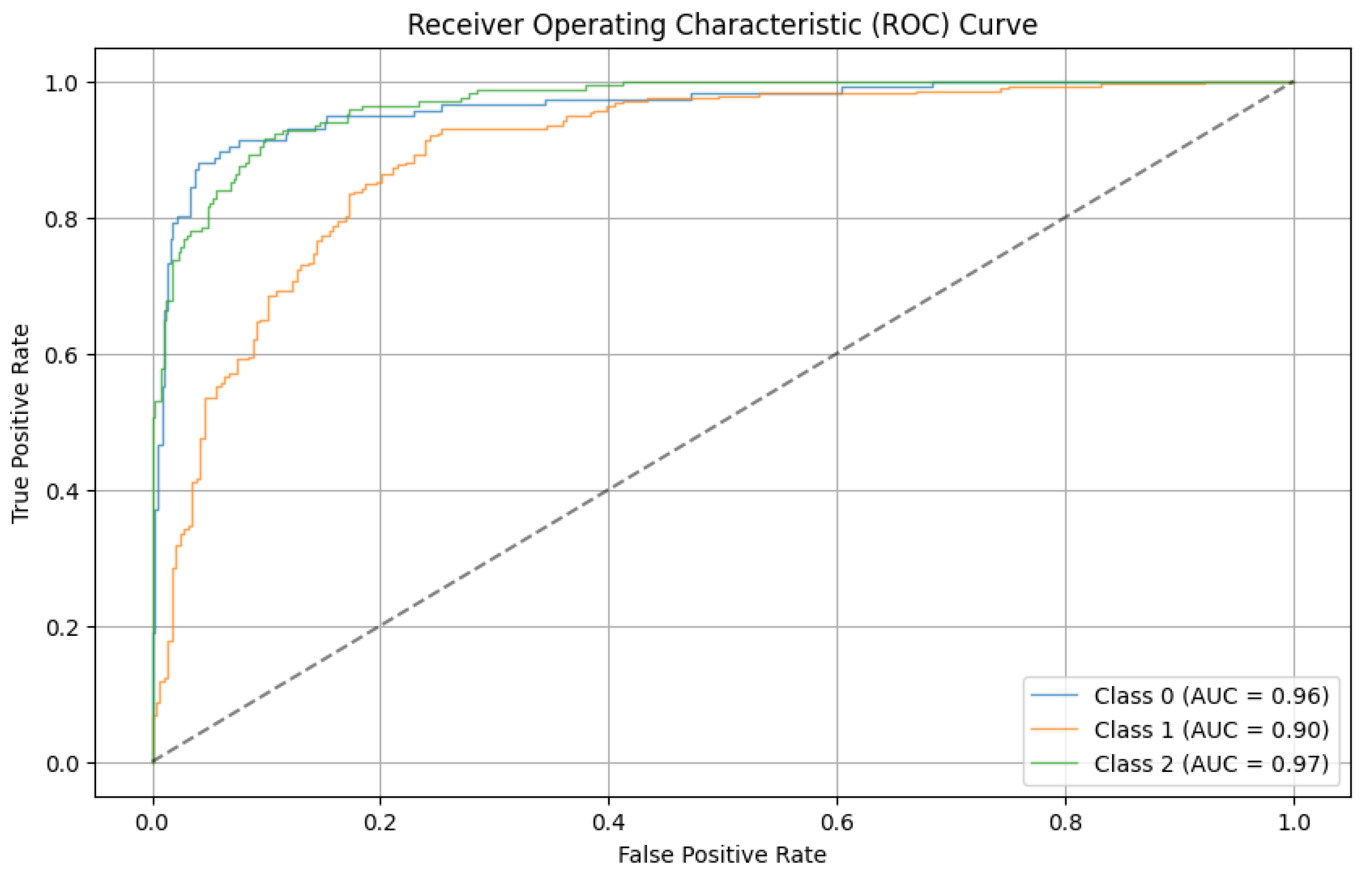
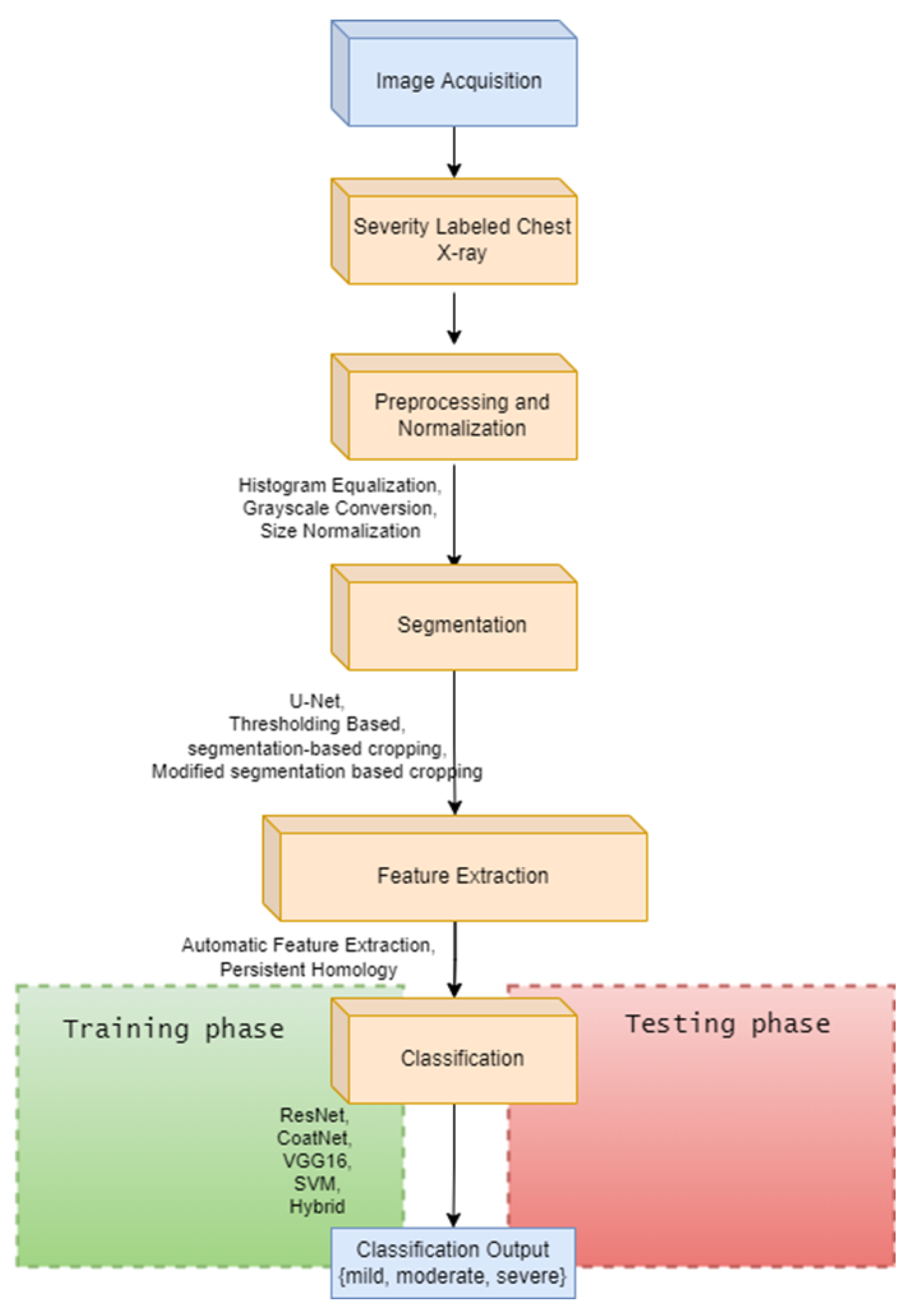
The development of a severity classification system for patients with COVID-19 via chest X-ray was the focus of this study. To improve the classification performance, we introduced a hybrid of convolution, self-attention, and persistent homology feature extraction techniques.
Histogram equalization is applied to enhance the quality of the images. The dataset was then normalized to have an identical size, color, and storage format. Thresholding, deep learning, and segmentation-based cropping techniques were compared. Next, the automatic feature extraction techniques ResNet-50, VGG-16, and CoatNet are studied along with the persistent feature extraction technique. A hybrid of persistent homology and the best automatic feature extractor was studied.
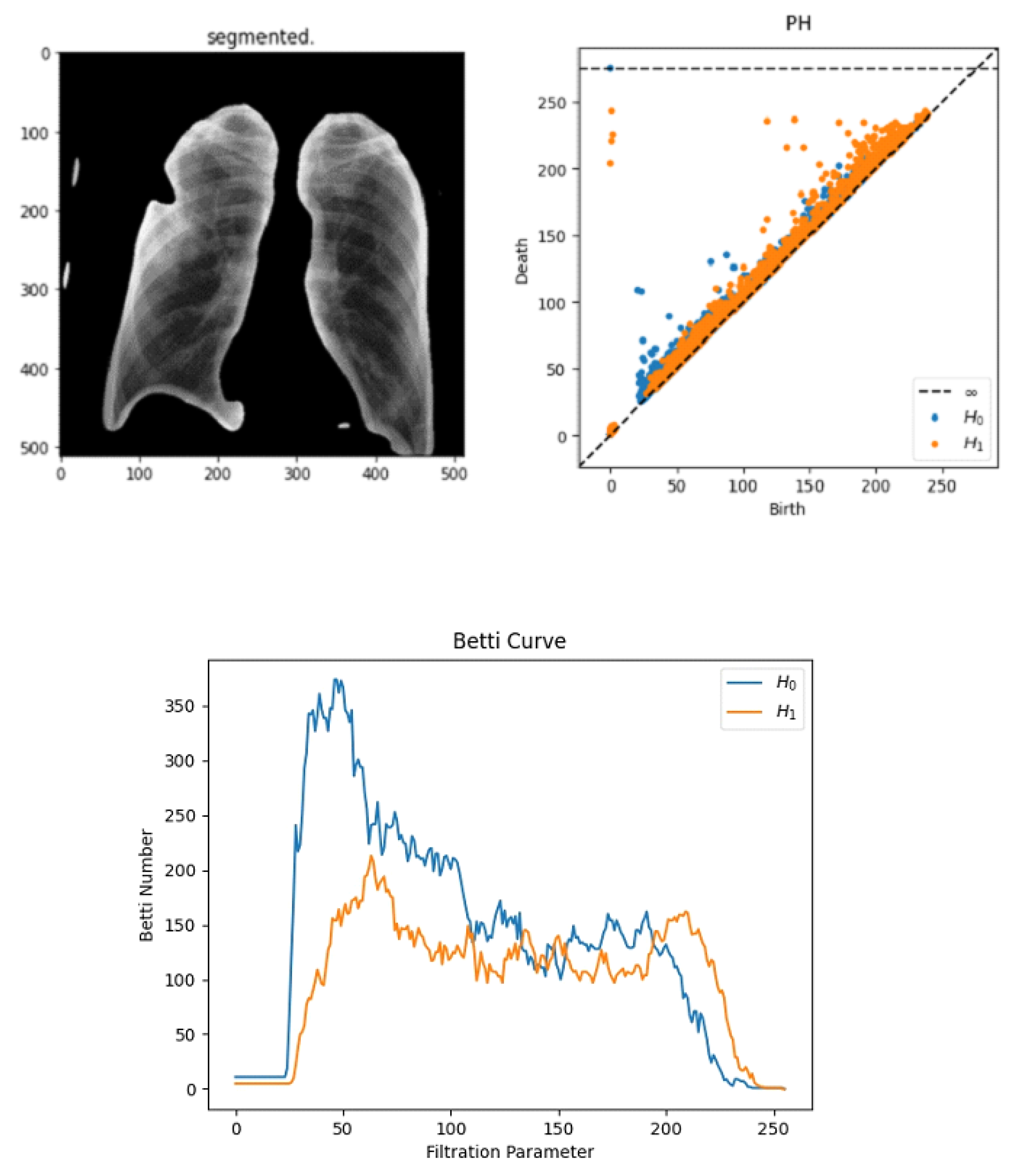
We find that using the modified segmentation-based cropping technique does a good job of containing the lung regions while eliminating unnecessary regions. From the feature extraction technique, we find that persistent homology vectorized as Betti curve features contributes to the overall accuracy. However, the amount of improvement is not large. The reason for this could be that the Betti number (curve) representation of persistent homology is unstable. The proposed model outperforms previously used CNN models (ResNet-50 and VGG16) as well as the final SVM classifier. This is because the proposed model takes advantage of the complementary strengths of each component:
Local Spatial Feature Extraction and Translation Equivariance: CNNs excel in extracting local spatial features and textures, crucial for identifying anatomical landmarks and texture anomalies in medical images. Their translation equivariance allows for effective generalization across varying anatomical positions, a common challenge in medical applications [
37].
Global Context and Long-range Dependencies: Vision Transformers, through self-attention mechanisms, capture global contextual relationships within images, effectively modeling correlations between distant regions. This capability is particularly beneficial for detecting disease signs that may be spread across different parts of the lungs, which can be challenging for purely CNN-based methods [
13].
Topological Feature Extraction: Persistent homology provides the ability to capture global structural and topological characteristics, including connectivity, holes, and voids, which are not adequately addressed by CNNs or transformers alone. These topological features offer robust descriptors that remain informative despite variations in local pixel intensity or minor spatial perturbations, enhancing discriminative power in clinical imaging tasks [
15,
39].
In contrast to other architectures, such as attention-gated CNNs and transformer-only architectures, our hybrid approach offers a more comprehensive analysis. Attention-gated CNNs primarily focus on local attention within convolutional feature maps, which may limit their ability to capture global relational information compared with transformers [
40]. Transformer-only architectures, while proficient at global context modeling, may struggle with local image structure and require substantial training data to effectively learn localized features [
41]. Our approach compensates for these limitations by integrating convolutional layers that efficiently handle local features, thereby creating a robust framework that leverages the strengths of each component.
The severity classification of COVID-19-infected lungs is different from the diagnosis problem, as there is no ground truth for verifying the class. In the case of diagnosis, there are verifiable ground truth tests such as RT–PCR (Reverse Transcription Polymerase Chain Reaction). The lack of such reliable tests makes developing a reliable dataset difficult. For severity classification, human radiologists must label chest X-rays manually. The problem with this approach is the fact that human radiologists subjectively declare the severity of cases. Subjective assessments by radiologists are known to have lower sensitivity because of the difficulty of visual interpretation of signs of disease on chest X-ray [
42]. This explains why the performance of the model is not significantly higher (than 83%). However, an improvement could still be made by choosing a more stable representation of homology features with the addition of qualitative information about the features as well (orientation and location, for example). Additional improvements could have been made by including other clinical information in addition to the chest X-ray images.
While our hybrid approach demonstrates significant advantages in classifying COVID-19 severity, it is essential to acknowledge certain limitations and potential areas for further improvement.
Segmentation Techniques: Each segmentation method evaluated presents distinct limitations. Thresholding-based segmentation methods are notably sensitive to image contrast and lighting variations, potentially affecting accuracy in images with severe pathological changes or varying imaging conditions [
23]. Conversely, deep learning-based approaches such as U-Net, despite offering detailed segmentation maps, rely heavily on large amounts of precisely annotated training data and may perform suboptimally when encountering previously unseen pathological manifestations [
24]. To enhance robustness, integrating ensemble or adaptive segmentation methods that dynamically adapt to different image conditions could be beneficial [
30].
Persistent Homology (Topological Features): The use of Betti curves in persistent homology presents inherent instability, particularly under slight variations in data, posing challenges to reliable clinical implementation [
33]. To address this, alternative stable representations such as persistence landscapes or persistence images have been proposed and validated in the literature, offering robustness to minor data perturbations and potentially enhancing classification reliability [
16]. Future research should explore these stable topological representations to further improve model performance.
Deep Learning Classifiers (CNNs and Vision Transformers): Convolutional neural networks (CNNs), while powerful in extracting local spatial patterns, inherently lack extensive global contextual understanding due to their limited receptive fields [
18]. Vision Transformers (ViTs), although capable of modeling global interactions effectively, require substantial datasets and might not capture detailed local patterns adequately [
13]. Combining CNNs and ViTs within hybrid architectures like CoatNet leverages the strengths of both local and global feature extraction, yet introduces complexity and computational overhead. Future studies should incorporate detailed interpretability analyses, employing methods such as SHAP and Grad-CAM, to elucidate clearly the specific contributions of CNN-based and Transformer-based features [
43,
44].
Hybrid Model Considerations: Our proposed hybrid model, combining convolutional, self-attention, and topological methods, naturally faces interpretability challenges due to its complexity. To address this, detailed interpretability frameworks and rigorous sensitivity analyses should be integrated into future research to validate the model’s clinical applicability comprehensively. Moreover, comparative studies against simpler fusion architectures, such as attention-gated CNNs or purely transformer-based models, are recommended to highlight clearly the incremental advantages of our proposed method.
Behavior Under Different Conditions: The model’s effectiveness could vary significantly under different imaging conditions and levels of severity. In particular, severe cases or lower-quality images may pose challenges due to indistinct pathological boundaries. Future work could explore model performance across varying image qualities and acquisition settings to ensure robustness and generalizability in real-world clinical scenarios.
6. Conclusions
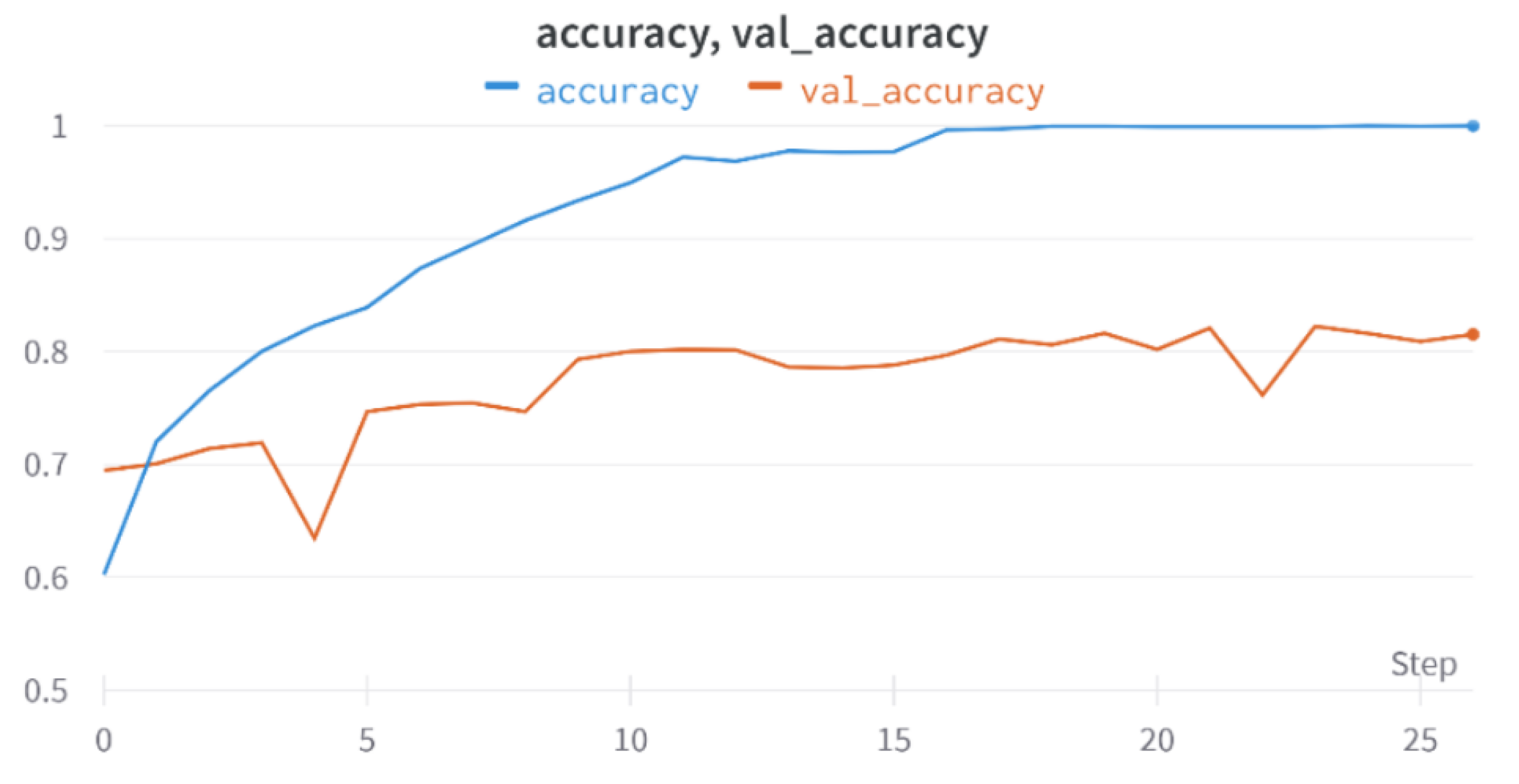
In conclusion, effective testing and patient monitoring are crucial in the fight against the pandemic, and imaging techniques such as X-ray and CT scans of the lungs play a vital role in diagnosing and assessing the severity of patients. X-ray imagery is faster, more cost-effective, and widely available, making it an important tool. Machine learning models based on chest X-ray images can assist medical professionals in quickly evaluating the severity of COVID-19 cases. Previous attempts using convolutional neural networks (CNNs) have limitations, but Vision Transformer networks offer improved capabilities for accurately analyzing input images. Topological data analysis techniques can extract important features from chest X-ray images. In this work, a combined network incorporating convolution, self-attention, and persistent homology (TDA) was trained and tested on a labeled dataset. The proposed method outperformed previous approaches, with an accuracy of 82.23% and an average sensitivity of 94.65%. Compared with other segmentation techniques, the cropping technique for lung segmentation yielded better results. Further analysis revealed slight advancements, and applying any segmentation technique improved the classification accuracy by 1.66%. The hybrid model, which incorporates topological features, achieved an additional 1.73% improvement. Future work could explore more stable persistent homology techniques and ensemble models with CNNs. Providing additional input data, such as vital signs, could enhance clarity and reduce subjectivity in the classification of X-ray images.