1. Introduction
Assisted reproductive technology (ART) is a treatment for individuals or couples, who are unable to conceive a child. ART techniques such as in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) involve fertilization of the egg outside the female body. Several oocytes (unfertilized eggs) are surgically removed from the ovary of the woman and fertilized with sperm in a laboratory, resulting in embryos. The embryos are cultured in an incubator with optimal conditions for a maximum of five days. The cultured embryos can be transferred into the uterus, cryopreserved for subsequent transfers, or discarded. Typically, the embryo with the highest quality is transferred back to the woman’s uterus. The process of estimating the quality of each embryo and ranking the available embryos within a cohort is called embryo evaluation [
1,
2]. The embryo evaluation is carried out manually by embryologists. The embryologists rank each embryo based on various criteria proven to be correlated with successful implantation or childbirth [
3,
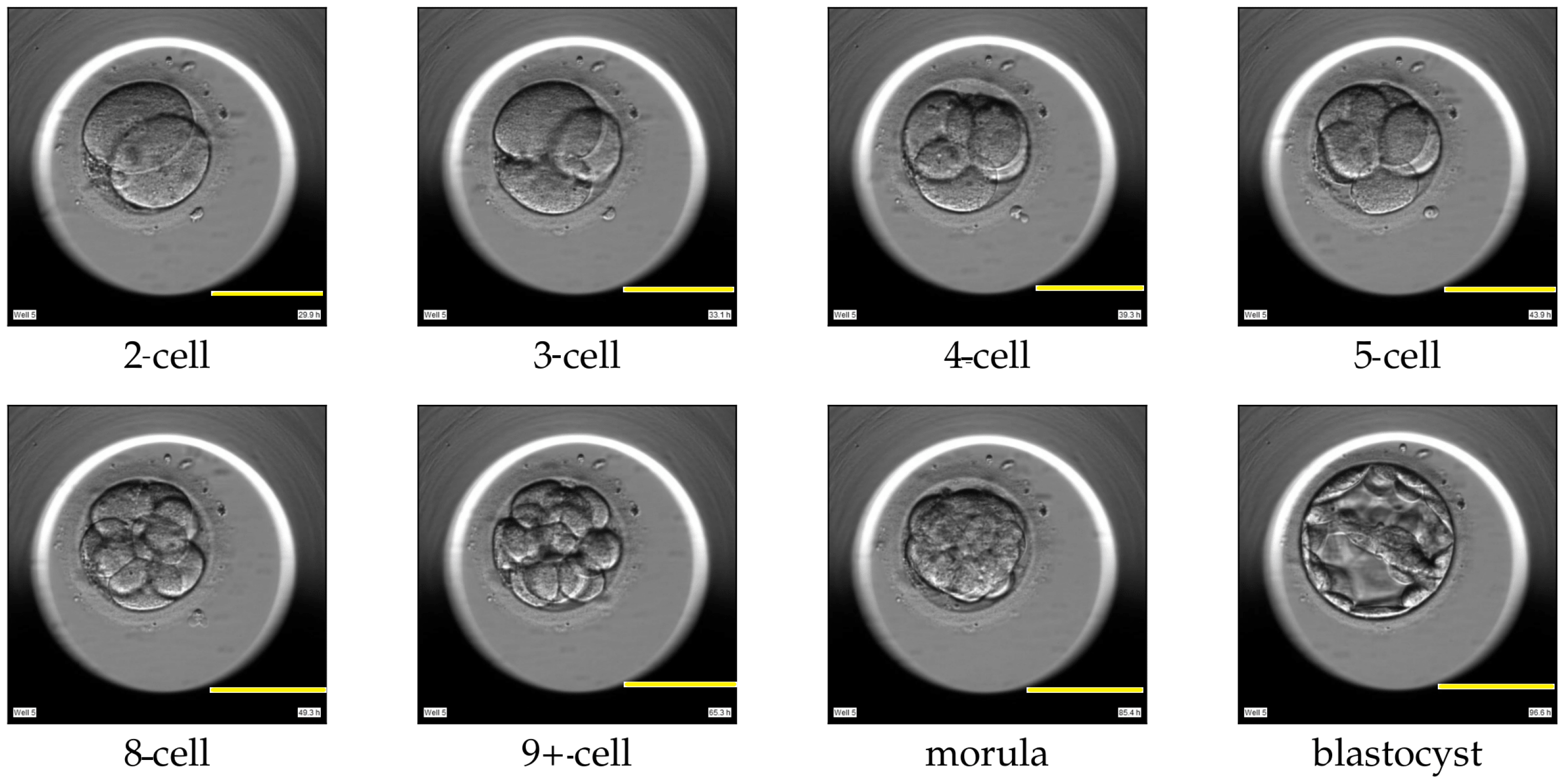
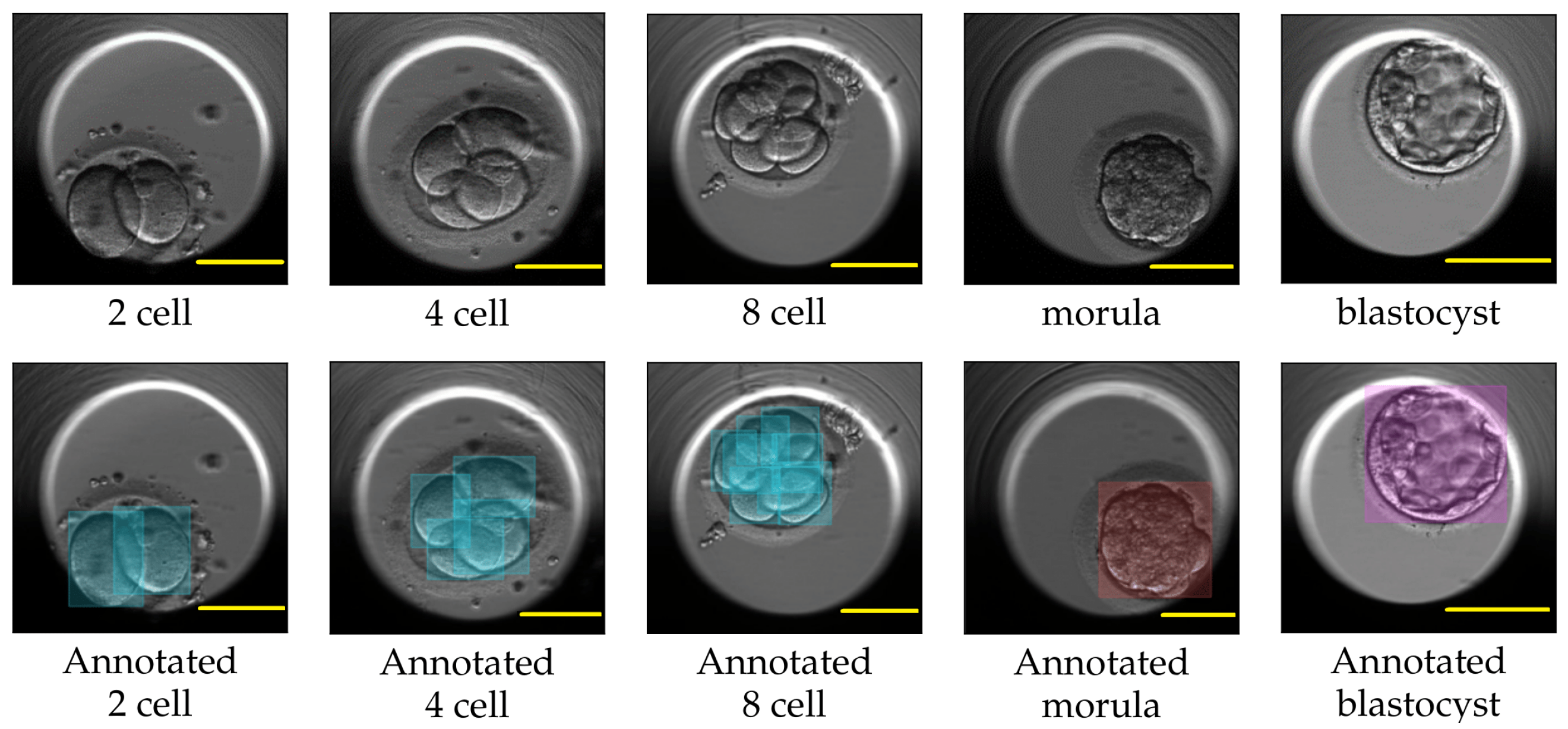
4]. One such criterion involves analyzing the dynamics of the cell cleavage stages, also referred to as the morphokinetic parameters related to embryo development. A cell cleavage stage is characterized by the number of embryonic cells and their subsequent cell division. An example of a human embryo monitored for development from the two-cell stage, three-cell stage, and four-cell stage to later stages such as the morula and blastocyst stage are shown in
Figure 1. The morula and blastocyst stages have a distinct morphology as compared to the early cell cleavage stages of embryo development. The morula stage is a compacted structure made of small-sized cells followed by a blastocyst, which is composed of hundreds of cells characterized into distinct features.
Evaluating the morphological development of cell cleavages is a relevant step in selecting embryos with high quality and viability [
5,
6]. The embryologists assess morphokinetic parameters such as changes in the cell morphology and the transitions during cell division to identify embryos with a higher potential to implant [
7,
8]. The information on cell cleavage duration is important for embryo evaluation [
9]. The time between subsequent embryo cell divisions, including both absolute and relative timings is relevant [
10]. Research has shown that in the embryos with a higher potential to implant, the cleavage from two cells to eight cells occurred comparatively earlier [
11] than in embryos that were unable to implant. A study concluded that evaluating the exact timing of embryo division in the early cleavage stages has a high potential to predict embryo quality [
12]. Another study revealed that the time taken to divide to five cells and the time between the division from three cells to four cells can effectively determine the embryo quality [
13]. Indeed, the evaluation of the exact timing of early events in embryo development is a promising tool for predicting embryo quality [
12], and progression from one cleavage to the next cleavage represents a noninvasive marker of embryo development potential [
14,
15]. Therefore, detecting the cell cleavage stages and the timings of successive cleavage during the preimplantation phase can provide valuable insights into embryo viability.
Usually, embryologists manually examine the cell cleavage stages and the length of the cleavage cycles [
16], and it should take less than 2 min to annotate a single embryo, but often a single patient has multiple embryos (5–10 embryos), so it can take up to 20 min [
17]. However, this task can be tedious and prone to subjectivity. The process can be automated using artificial intelligence (AI) or, specifically, object detection algorithms and optical character recognition (OCR). In fact, in recent times, several AI algorithms have been applied to automate embryo evaluation [
2,
18,
19,
20]. The application of object detection algorithms for medical image processing is common [
21]. The medical domain images usually have the object of interest as a small surface area and blurred boundary [
22]. However, the algorithms effectively detect the object in both images [
23] and video stream [
24]. A similar characteristic was observed for embryo cells with the application of time-lapse imaging. The time-lapse technology (TLT) enables the continuous monitoring of embryo development [
25,
26] and provides comprehensive information regarding the morphokinetics of embryo development including observations for events such as cell division or the transition between different cell cleavage stages [
27]. With the application of an object detection algorithm, we can detect cells inside an embryo and determine the associated cell cleavage stage. The time-lapse imaging also captures the elapsed time since the start of fertilization. Depending upon the time-lapse system software, usually, the hours post insemination (hpi) is appended to each frame of video and can be read by the application of optical character recognition (OCR). Thus, using object detection and OCR we can automate the process of finding the start of an observed cell cleavage stage and recognizing the associated time in hpi (absolute or relative) for the stage. The automation will benefit embryologists in their daily tasks and can be used as a decision support system.
The software iDAScore from Vitrolife automatically estimates cell division events in time lapse. The feature is referred to as Guided Annotation [
28]. The software requires a license and also the presence of Vitrolife EmbryoScope, EmbryoViewer software, which makes it cost intensive. Moreover, the quality of the TLT video is important for Guided Annotation’s efficient performance. For example, during the entire period, the embryo should be centered and in focus with entire embryo region being visible. There are alternative noninvasive approaches to count the number of cells and their location in time-lapse images of embryo development [
29,
30,
31]. Their algorithms are based on computer vision and machine learning and predict only the cell count but no time annotations for the cell cleavage stages. Moreover, the algorithms require additional preprocessing steps such as image processing, detecting the embryo location, or extra filters to grasp the morphology progression along the temporal domain. This makes the approaches resource-demanding. Furthermore, the approaches only detect the early stages of embryo development and do not apply to later stages such as morula and blastocyst.
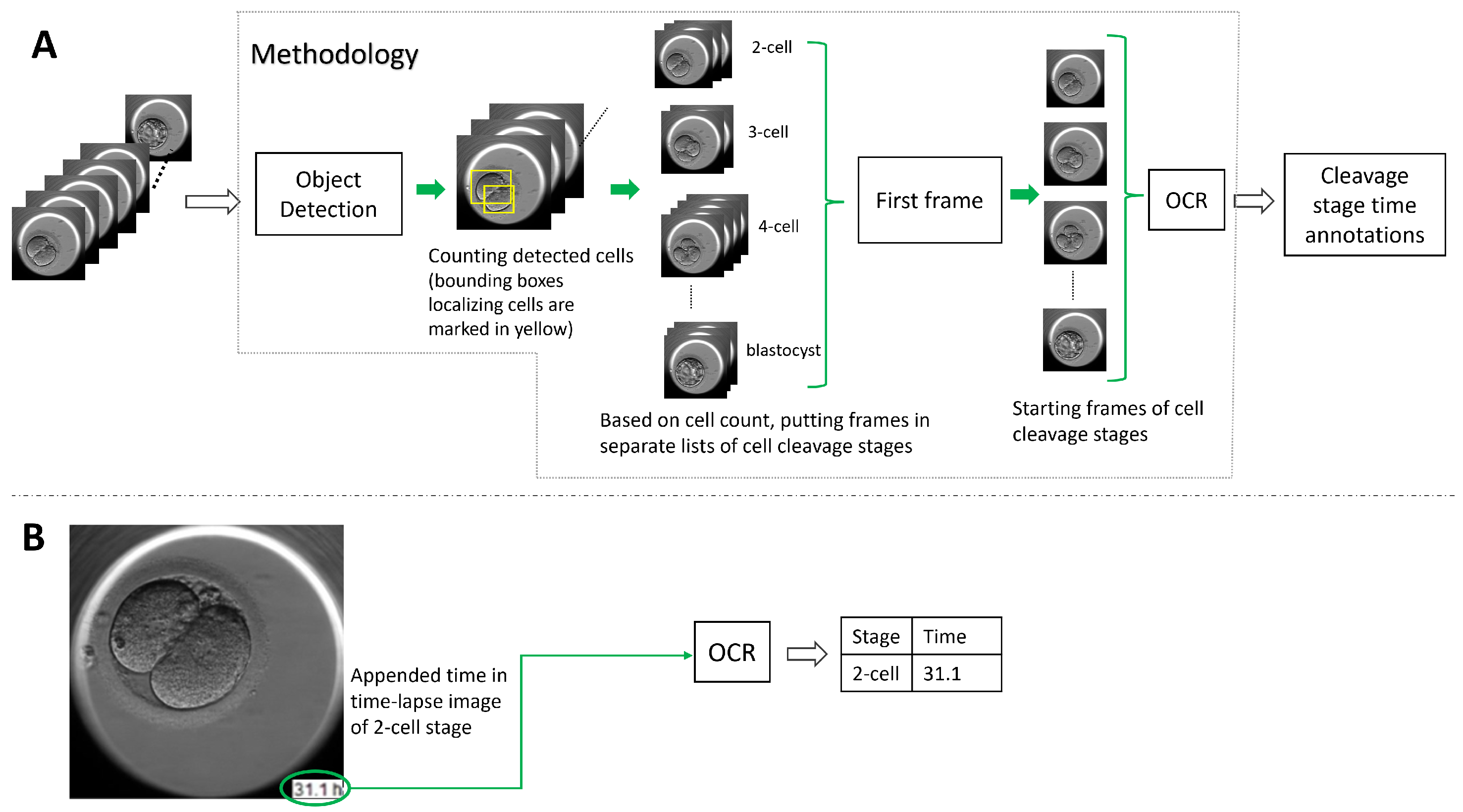
To address the lack of time annotation in previous research, in this article, we suggest a novel methodology for identifying the start and duration of cell cleavage stages in TLT videos. The methodology uses object detection algorithms to analyze each frame in the video and identify instances of cells, as well as the presence of the morula and blastocyst stages. The frames that belong to a cell cleavage stage are grouped together based on the number of cells or the presence of the morula and blastocyst. The first frame in each group is annotated as the start of the cleavage stage, and the hpi present on that frame is read using OCR. If the hpi value is missing, the methodology derives it using the video frame rate and time-lapse system configuration. We evaluated two object detection algorithms (YOLO v5 and DETR, explained in
Section 3.1) and three OCR techniques (pytesseract, EasyOCR, and Keras-OCR, explained in
Section 3.2) with our methodology. In
Figure 2, we outline the methodology’s workflow.
The suggested methodology is capable of not only locating the cells but also predicting the start of all cell cleavage stages observed in a TLT video. Furthermore, we found that the presence of artifacts or fragmentation in the datasets did not affect the performance, since our methodology was able to differentiate them from the cells. The methodology operates directly on TLT video frames without any additional preprocessing steps or software license installation, making it both cost- and time-effective. For patients with the number of embryos in the range of 5–10, the methodology used, on average, around 3 min to compute the annotations of all the TLT videos. To best of our knowledge, besides iDAScore, there exists no other software tool that computes automated cell cleavage annotations. Thus, the combination of object detection with OCR in the field of ART and for the automated annotating of morphokinetics parameters is novel.
The main contributions in the article are as follows:
We have developed a fully automated framework for tracking cell divisions and counting cells in embryos.
Our methodology predicts and annotates the start time of cell cleavages without the need for any manual intervention by embryologists.
Our methodology effectively detects the starting frame of cell cleavage stages from one cell to five cells, and achieved an F1-score of 0.63 and an accuracy of 0.69. For detecting the starting frames for stages with cell counts greater than five, our methodology was delayed by 30–32 frames on average, considering videos with a frame rate of eight frames per hour. The time annotations for the two-cell to five-cell stages were delayed by 2–3 hpi on average.
Our methodology’s versatility allows it to accurately detect not only the number of cells in an embryo but also the distinct morphological structures of later cell cleavage stages, such as the morula and blastocyst.
We examined the relation between the level of fragmentation within an embryo and the performance of our proposed methodology. Embryologists’ validation confirmed that our approach did not confuse cells with small-sized fragments.
Our methodology computes start-time annotations (in hpi) for videos in real time, making it suitable for clinical applications. The shortest video was annotated in 26 s, while the longest video took approximately one minute.
The rest of the paper is organized as follows:
Section 2 provides an overview of the existing methodologies automating the detection of the cell cleavage stages, and
Section 3 provides an overview of the state-of-the-art object detection algorithms and OCR libraries.
Section 3 also describes the principle theory behind time-lapse systems.
Section 4 describes the data used for training and evaluation of the methodology, and
Section 5 provides an overview and discusses various components of the methodology.
Section 6 and
Section 7 discuss the results and their limitations along with suggestions for future research. Finally,
Section 8 concludes the paper and highlights the main findings.
4. Data
The dataset consisted of TLT videos collected retrospectively by embryologists working at the Fertilitetssenteret. The Fertilitetssenteret is a fertility clinic in Oslo, Norway. The embryos were cultured inside an EmbryoscopeTM with similar time-lapse imaging specifications as described in
Section 3.3. The dataset contained two types of videos: embryos that had been transferred to females and embryos that had been frozen or cryopreserved for later use. We used two separate categories to potentially improve the generalizability of the suggested methodology. For example, the frozen videos had a higher rate of fragmentation compared to the transferred videos. We used the transferred videos to train and validate the object detection algorithms, which we explain in
Section 5.1. The dataset consisted of 250 videos and is referred to as the ‘TransferV’ dataset in the rest of the paper. Furthermore, the TransferV was further divided into a training set and an evaluation set consisting of 200 and 50 videos, respectively. The training set was used for training the object detection algorithms to locate instances of cells, morula, or blastocyst, while the evaluation set was used for evaluating the object detection algorithms and tuning the hyperparameters for efficient detection of the cell cleavage stages.
While TransferV was used to train and evaluate the object detection algorithms, the frozen embryo videos were used to evaluate the performance of our methodology to predict the start of the cell cleavage stages. The dataset consisted of 29 videos and is referred to as ‘FrozenV’ in the rest of the paper.
Table 1 summarizes the different datasets that were used in this study.
For each video in the TransferV dataset, the embryologists at the Fertilitetssenteret reviewed all the video frames and manually annotated the start (hpi and frame number) of the observed cell cleavage stage. We used the annotations to extract the representative frame by marking the start of the cell cleavage stages in the videos. The training set consisted of frames only belonging to the two-cell, four-cell, five-cell, eight-cell, nine+-cell, morula, and blastocyst cleavage stages, while the evaluation set consisted of representative frames for all the cell cleavage stages from the one-cell to the blastocyst stage. The frames in training set were annotated with class labels as cells, morula, or blastocyst using the applications Labelbox [
47] and Roboflow [
48].
Figure 8 shows some labels created with LabelBox. We further applied augmentation techniques, such as horizontal and vertical flip and rotation between −30° amd 30°, to increase the number of class labels. In total, we created 3868 class labels, 3490 for cells, 188 for morula, and 190 for blastocysts. The accurateness of the created class labels using LabelBox and Roboflow were approved by the embryologists.
Three embryologists independently reviewed the FrozenV dataset videos frame by frame and annotated the start of the cell cleavage stages (time and frame number). These embryologists are referred to as E1, E2, and E3 in the rest of the paper. The annotations from all three embryologists were used to evaluate the methodology’s performance in predicting the start of cell cleavage stages. However, 0.5 percent of the manual annotations were incorrect. To rectify this, we replaced the incorrect annotations with the average value calculated from the annotations of the two other embryologists.
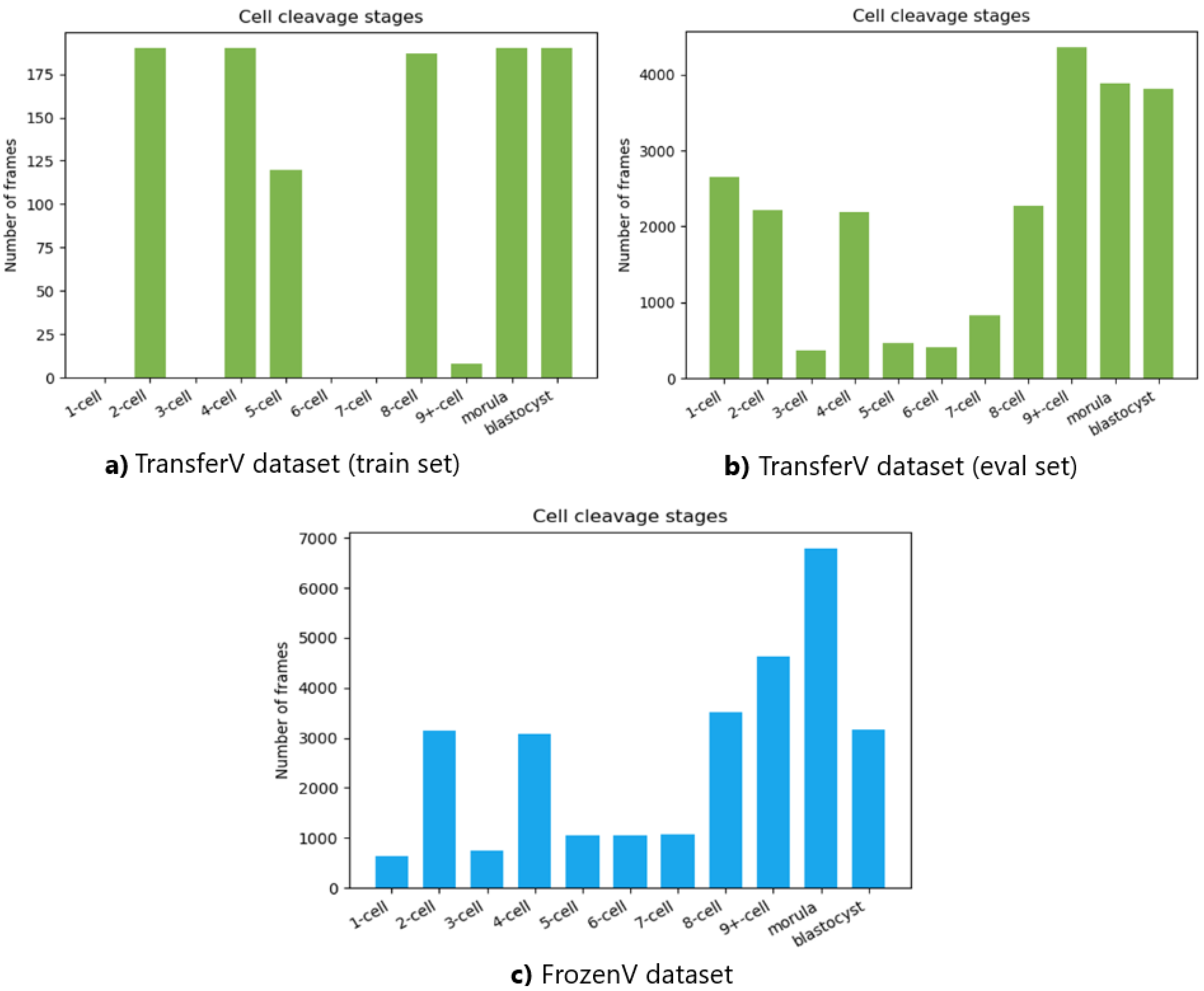
The number of frames in the videos for each cell cleavage stage in the TransferV and FrozenV datasets is shown in
Figure 9. The cell cleavage stages are characterized by the presence of a specific number of cells, morula, or blastocyst. Thus, we also present the distribution of instances for cells, morula, and blastocysts for both datasets in
Table 2.
As mentioned before, E1, E2, and E3 independently annotated the FrozenV dataset. Thus, we combined the three independent set of annotations using majority vote and represented our best estimate for the ground truth.
Table 3 shows the portion of frames where the different embryologists agreed with the majority vote, for the different cleavage stages. We see that for the two-cell to four-cell stages, the agreement was quite high, but from the five-cell stage the level of disagreement increased.
5. Cell Cleavage Detection
In this section, we describe the suggested methodology to effectively detect the start of the cell cleavage stages. The methodology involved identifying objects such as cells, morula, and blastocysts in a frame and counting the number of detected objects. The object detection was further used to assign each frame to the corresponding cell cleavage stage and annotate the time in hpi for the start of these cleavage stages. In
Section 5.1, the training of the object detection algorithms locating cells, morula, and blastocyst in embryo images is explained. In
Section 5.2, the methodology to detect the cell cleavage stages based on the output from the trained object detection algorithms is explained. Finally, in
Section 5.3, the methodology to annotate the start time of the cleavage stages in the hpi is explained. In
Section 5.2, we explain the tuning of the methodology’s hyperparameters, the central part of the suggested approach in predicting the start of the cell cleavage stages. Both
Section 5.1 and
Section 5.3 build on
Section 5.2, which represents the main idea of the methodology.
5.1. Object Detection
The methodology uses object detection algorithms to detect and segment instances of objects: cells, morula, and blastocysts. Thus, in this Section we describe the training of YOLO v5 and DETR to detect the objects. Both the object detection algorithms were trained on the class labels created from the training set of the TransferV dataset (explained in
Section 5.1) The training set was divided into a 4:1 ratio for training and validation.
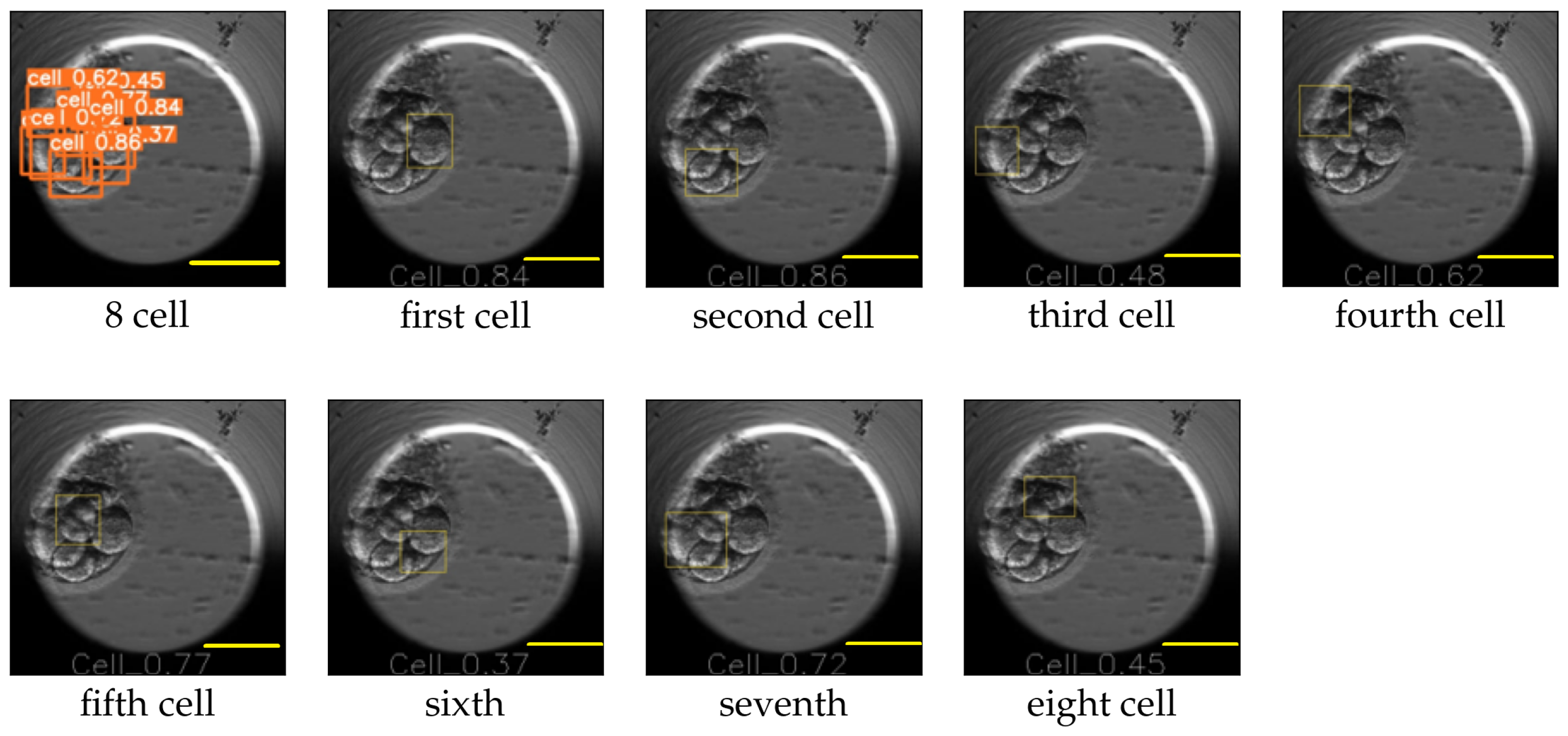
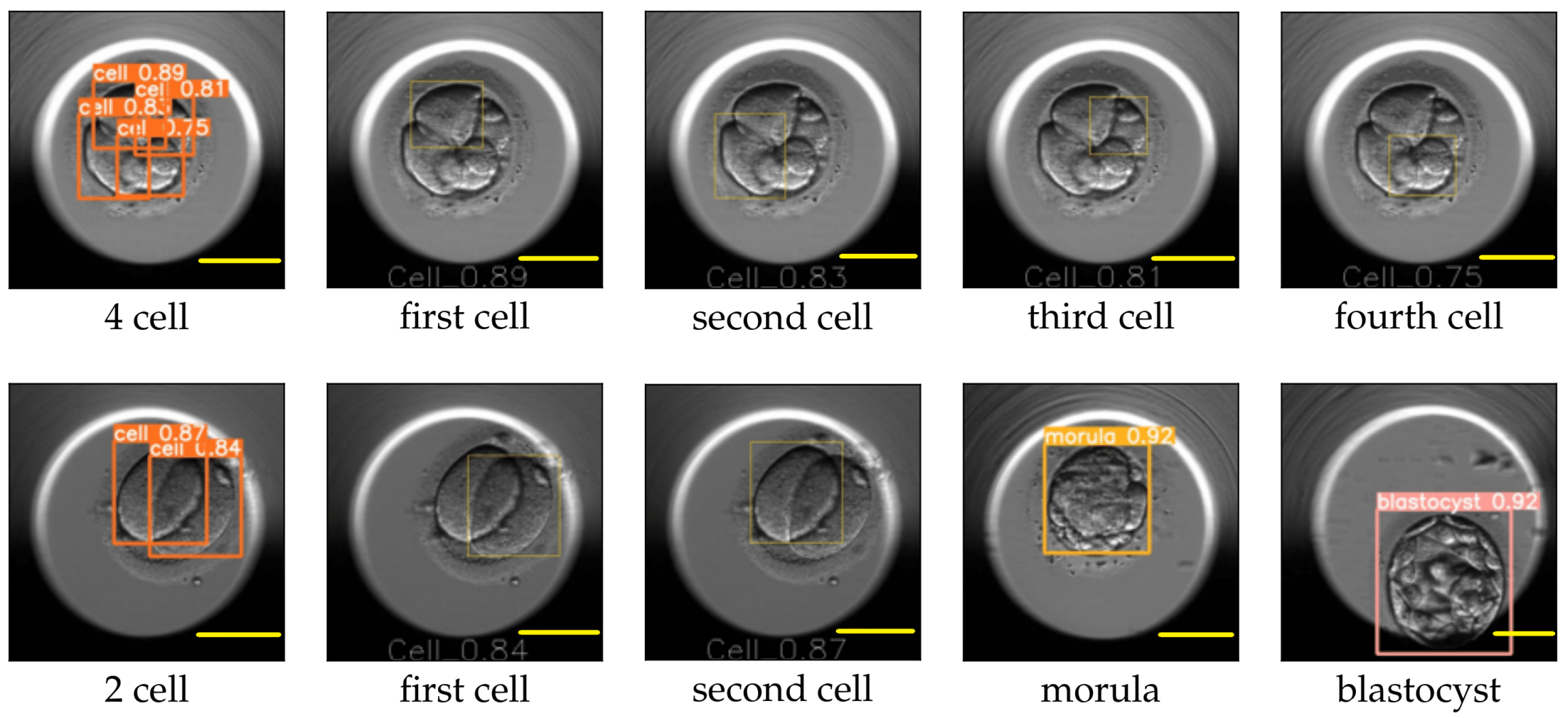
The image size used for training YOLO v5 was 416 × 416, and the value for the hyperparameter determining the minimum bounding box confidence score was 0.30. YOLO v5 reported the mAP for the cell, morula, and blastocyst equal to 0.65, 0.78, and 0.80, respectively. An example of the YOLO predictions for the eight-cell, four-cell, two-cell, morula, and blastocyst stages is explained in
Figure 10 and
Figure 11. The figures replicate the input image and draw each bounding box separately along with the class probability of the detection.
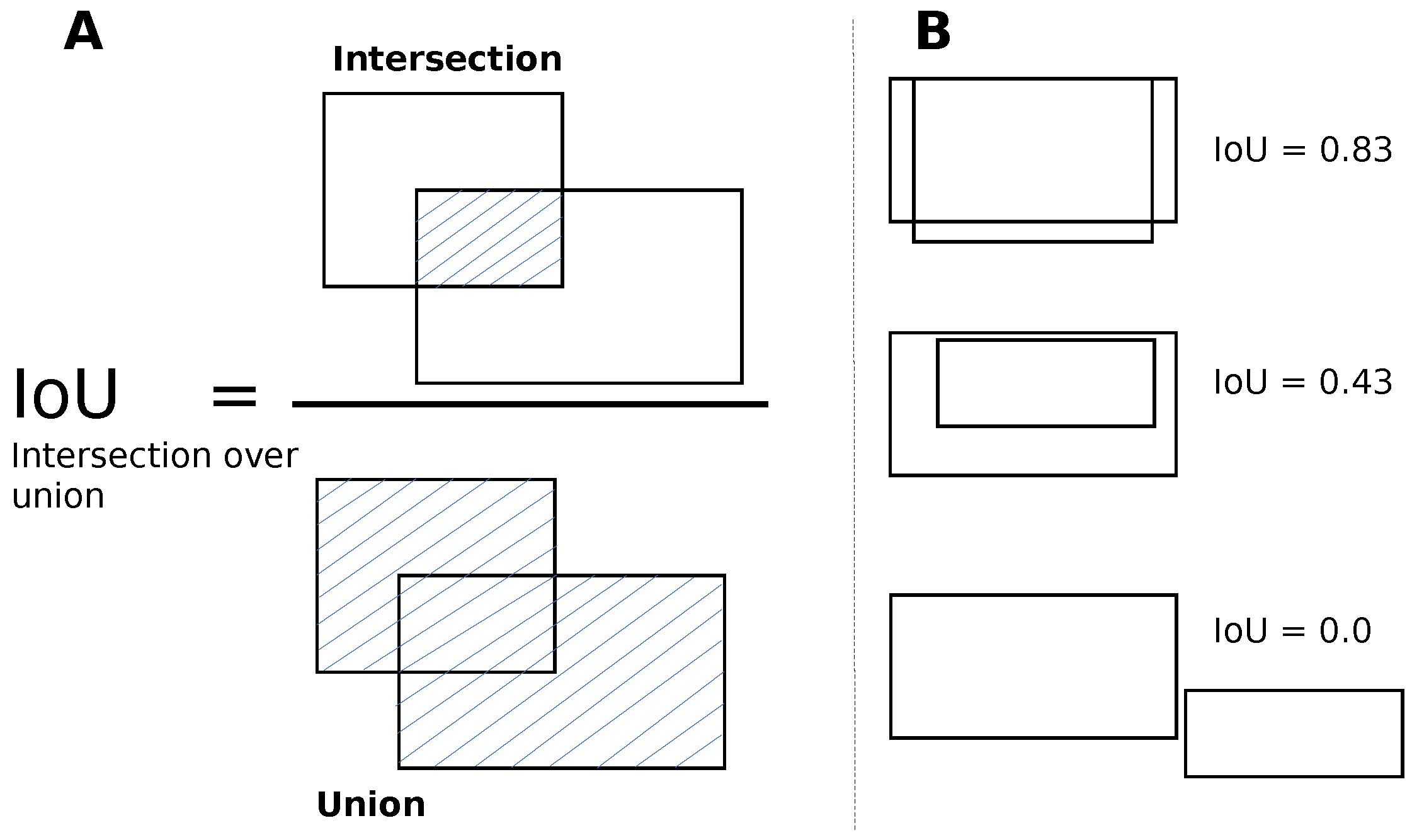
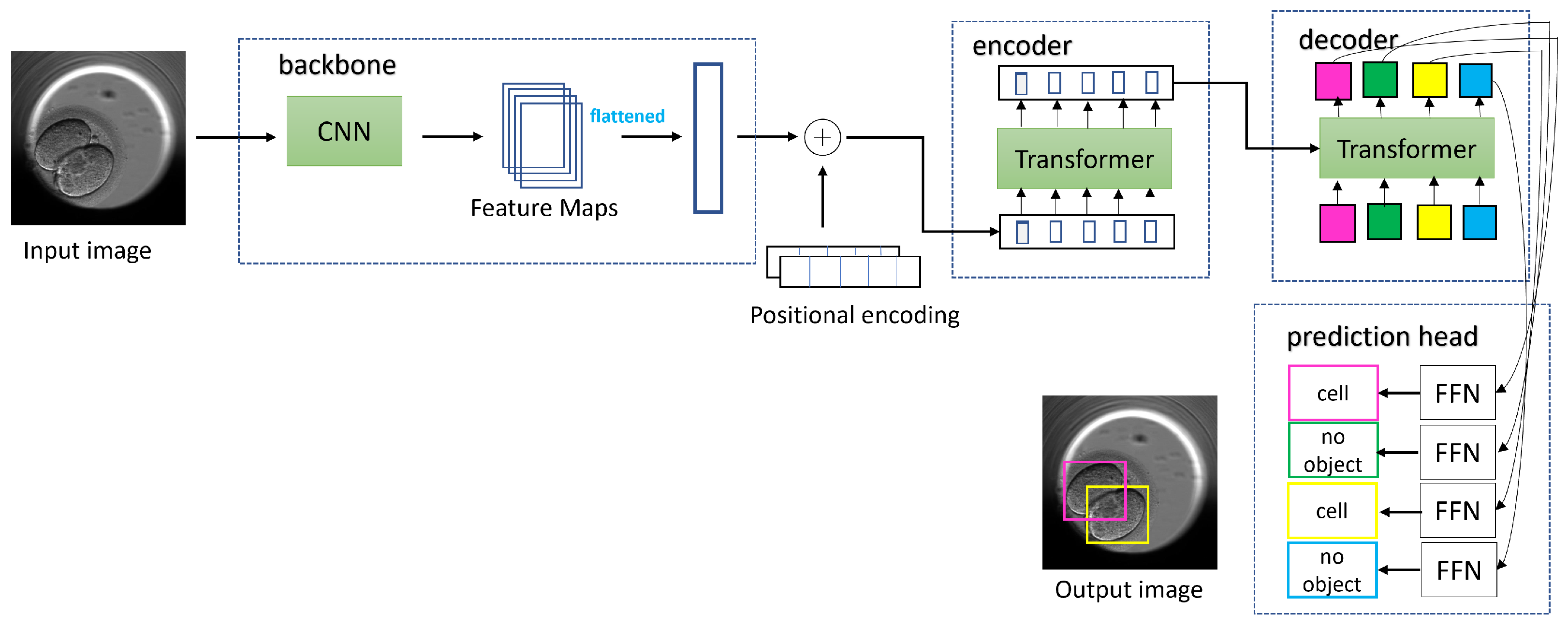
To train the DETR model, we followed a multistep process. Initially, we set the image size to 416 × 416 and the batch size to eight. During training, we updated the gradients after every four batches, which was equivalent to 32 images. We replaced the last layer of the model (FFN) with a custom layer that predicted the bounding boxes and class labels for the cells, morula, and blastocysts. In the first ten epochs of training, we froze the backbone (ResNet-50) and the transformer (encoder, decoder) while only training the last layer with a learning rate of 1 × 10
−3.In the subsequent 50 epochs, we unfroze the transformer and the last layer and only froze the backbone. We continued to train the last layer with the transformer, with learning rates of 1 × 10
−3 and 1 × 10
−4, respectively. After 60 epochs of training, we achieved an IoU validation loss equal to 0.22, an L1 loss equal to 0.05, and a cross-entropy loss equal to 0.16. For the next 170 epochs, we unfroze all the blocks, including the backbone, transformer, and last layer. We trained the model with learning rates of 1 × 10
−5, 1 × 10
−4, and 1 × 10
−3 for the backbone, transformer, and last layer, respectively. Finally, after the full training process, we achieved a final IoU validation loss equal to 0.16, an L1 loss equal to 0.04, and a cross-entropy loss equal to 0.14.
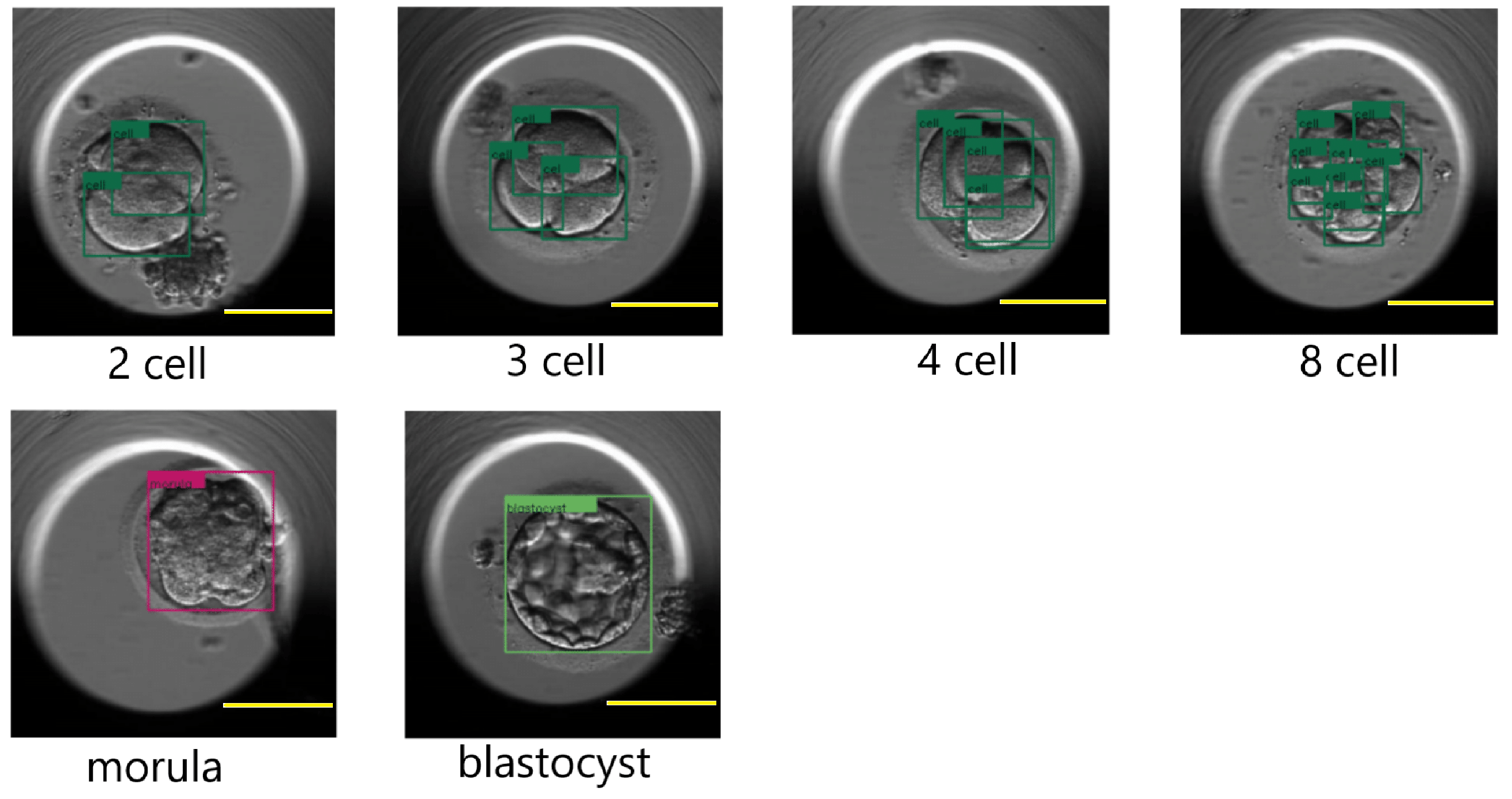
Figure 12 shows a few results of the DETR detecting cells, morula, and blastocysts.
5.2. Detecting the Start of Cleavage Stages
In this section, we focus on identifying the cell cleavage stages associated with the objects detected (cells, morula, or blastocyst) in the previous section. The trained object detection algorithms localize the objects in a frame and provide class probabilities for each detected object. The suggested methodology counts the number of detected objects in a frame and maps the frame to the corresponding cell cleavage stage if the class probability of each detected object is above a specified threshold value. The cell count and threshold values are parameters used for predicting the start of the cell cleavage stage. For example, if two cells were detected with each cell class probability above the specified threshold for the two-cell stage, the frame was classified as a two-cell stage.
The threshold values were determined empirically by evaluating the object detection algorithm on the evaluation set of the TransferV dataset. For each frame in the evaluation set, the embryologists evaluated the methodology’s performance and set the threshold values for each cell cleavage stage based on their analysis. The threshold values for the cell cleavage stages were: 0.80 for the two-cell, 0.70 for the three-cell, 0.65 for the four-cell, 0.60 for the five-cell, and 0.50 for the six-cell up to 9+-cell. The threshold value for the morula and blastocyst stages was set to 0.90.
In a video, there can be several frames associated with a cell cleavage stage. Therefore, the methodology organized the frames for each cell cleavage stage, and the first frame from the series was annotated as the start of the cell cleavage stage. The frames put in a series should have matching values for the parameters cell count and threshold. Whether the methodology used YOLO v5 or DETR for the object detection, the same annotation scheme was used. To potentially reduce the noise in the computed probabilities, we also explored using the moving average of the probabilities for three and five subsequent frames, and compared these with the threshold value. However, using the probabilities without averaging turned out to perform better.
5.3. Computing Annotations for the Cleavage Stages Starting Time in hpi
In the previous section, we explained how to find the frame associated with the start of a cleavage stage. In this section, we explain how to find the corresponding time in the hpi. The time corresponds to the occurrence of the captured embryo development in the video recorded in hours (post insemination) by the TLT system. There were two categories of TLT videos: the first one, where the hours were appended to the video frame (as shown in
Figure 2) and the second one, where the time entry was missing from the frames. The suggested methodology used the OCR libraries to detect the encoded time when the hpi value was present in the video frames. We evaluated the OCR libraries: Pytesseract, EasyOCR, and Keras-OCR. Recall the introduction to the libraries in
Section 3.2. However, for the TLT videos where the frames were not annotated with hpi, the methodology calculated the hours using the duration of the video, the start time of fertilization, the number of frames, and the frame rate of the TLT system.
6. Results
In this Section, we evaluate the performance of the suggested methodology for annotating the start of the cell cleavage stages, both in terms of the frame number and hours. We summarize the performance of the methodology in predicting the start of the cell cleavage stage in
Section 6.1 and evaluate the performance in annotating the start time measured in hpi for the observed cell cleavage stages in
Section 6.2.
The cell cleavage stages of the morula and blastocyst have unique substages in terms of morphology. As the object detection component of our method was only trained to detect the first substage, we did not evaluate the morula and blastocyst with the other cell cleavage stages. Nevertheless, our method effectively detected the first substages of the morula and blastocyst, and this was independently verified by three embryologists. In the rest of the paper, and refer to the suggested methodology to detect the start of a cleavage stage, when YOLO v5 and DETR were used for the object detection, respectively.
6.1. Detecting Cell Cleavage Stages
In this Section, we present the evaluation results for the methodology in predicting the starting frame of the cell cleavage stages. The predictions were evaluated on the FrozenV dataset. The performance of the suggested method was compared with the majority vote of the three embryologists, described in
Section 4.
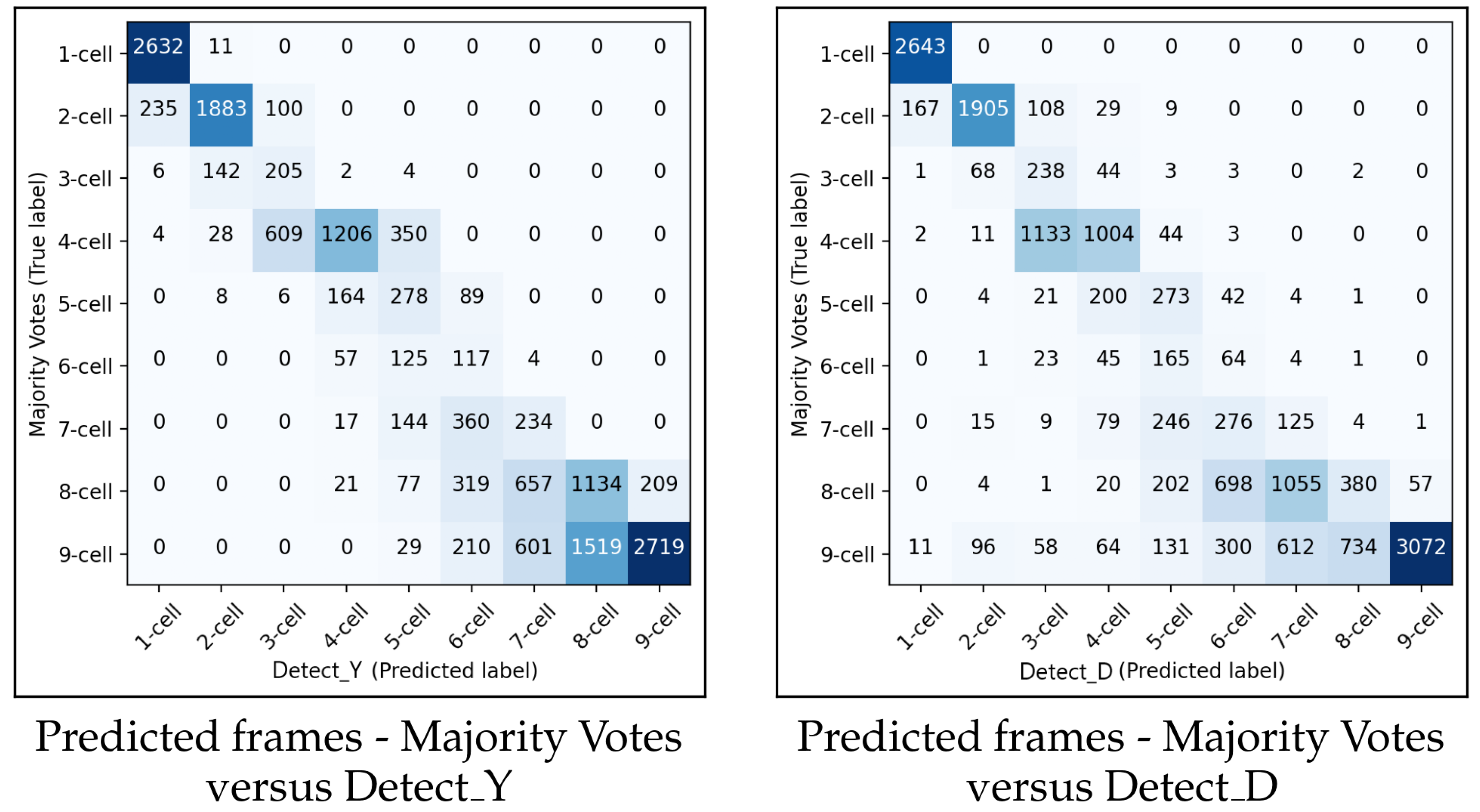
Figure 13 shows the confusion matrices for
and
for predicting the starting frame of a cleavage stage.
In general, the performances of and were similar. When the methodology made a wrong prediction, it was mainly with the adjacent cell cleavage stage. Additionally, in most cases when the methodology predicted incorrectly, it predicted too few cells. In the confusion matrix for , there were more misclassifications between the eight-cell stage and the adjacent stages of seven cells and nine+ cells compared to the previous cleavage stages. A similar pattern of misclassification was also observed in .
The methodology was further evaluated using the six performance metrics of precision, recall, specificity, accuracy, F1-score, and the Matthews correlation coefficient (MCC). Each metric evaluates the methodology’s performance on different parameters, providing a reliable and thorough analysis of its predictive capability [
49]. Precision quantifies the proportion of the correctly identified starts of the cleavage stages out of all the predicted starts of cleavage stages. Recall computes the ratio of the correctly predicted starts of a cleavage stage to all the predicted start frames indicated by the methodology. The F1-score is the weighted average of the precision and recall. For a cell cleavage stage, the specificity measures the proportion of predictions that were not mislabeled by the methodology as the start of another cell cleavage stage. The MCC metric provides an overall evaluation of the methodology’s accuracy in predicting the start of the cell cleavage stages and avoiding misclassifications with the start of the other cell cleavage stages. The MCC value ranges from −1 to 1, where a negative value indicates disagreement, a positive value indicates agreement, and a zero value indicates no agreement.
Table 4 shows the prediction performance of
v5 and
evaluated per cleavage stage.
The methodology performed the best for the one-cell and two-cell stages with consistent and high prediction rates. Specifically, the F1-score for the methodology using
was 0.95 and using
was 0.97 for the one-cell stage, and the F1-score value was 0.88 for the methodology using both
and
for the two-cell stage. Further, both the recall and precision values were high. The methodology demonstrated good prediction performance for the four-cell stage, with an F1-score of 0.66 using
and 0.55 using
. However, for the three-cell and five-cell stages, the methodology had lower discriminative ability and reported average recall and low precision values. These stages were also underrepresented compared to the other stages (FrozenV dataset in
Figure 9), which might have affected the performance. The methodology’s prediction performance dropped considerably for the six-cell stage and onward, with both recall and precision values decreasing. However, the methodology had high specificity values for all cell stages, indicating lower misclassification rates between different cell cleavage stages.
We conducted a performance analysis comparing the two object detection algorithms used in the methodology. had a higher MCC value of 0.58 compared to 0.53 for . Additionally, the accuracy rate was lower for compared to from the one-cell stages to the eight-cell stages. Thus, the methodology performed better using than using . Additionally, we evaluated the methodology using two other versions of YOLO: YOLO v5 with soft NMS and YOLO v7. However, these tests did not result in any improvement. In fact, the performance metrics reported by the methodology were lower than those obtained using from the five-cell stage onward.
Recall that
Table 3 shows the agreement rate between E1, E2, and E3 on the starting frame for cell cleavage stages. The overall agreement rate was much higher in comparison to the suggested methodology for predicting the starting frame. The average value for E1 was 0.90, E2 was 0.81, and E3 was 0.88. In comparison, the average accuracy rate for
was 0.57, and for
, it was 0.51. As mentioned in
Section 4, the observed fragmentation in the FrozenV dataset was higher than that of the TransferV dataset, but the change in the fragmentation rate had no consequence on the performance of
and
. The embryologists validated that the methodology did not mistakenly identify small-sized fragments as cells, even wjen the fragments had overlapping boundaries with the cells.
6.2. Annotating Hours Post Insemination for the Cell Cleavage Stages
In this section, we address the problem of predicting the time in hpi for the start of a cell cleavage stage. We used OCR to extract the timing information available in the video frames. In the previous section, we observed that performed better than and that could efficiently detect the frame marking the start of the cell cleavage stages up to five cells. Therefore, we evaluated the performance of in predicting the time in hpi up to five cells. We evaluated the OCR libraries pytesseract, EasyOCR, and Keras-OCR. We tested the OCR libraries such as pytesseract, EasyOCR, and Keras-OCR for digit recognition (time encoded in frames) and compared their performance against the time annotations obtained through majority voting. In recognizing the time digits present in the frames, pytesseract performed the best. The pytesseract library only confused the digits ‘4’ and ‘1’ in 0.2% of the tests. Both EasyOCR and Keras-OCR wrongly recognized digits as alphabets and with much higher percentages (EasyOCR: 4.8%, Keras-OCR: 3.6%). We also computed the time needed by the different OCR libraries as part of to annotate the whole FrozenV dataset. The annotation task was finished in 8.21 min with pytesseract, 9.09 min with EasyOCR, and 9.22 min with Keras-OCR.
Using with pytesseract, a time delay was observed in predicting the start of the cell cleavage stages compared to the embryologists’ majority votes. The average time delay for the one-cell stage was minimal, 1.24 hpi for the two-cell stage, 0.63 hpi for the three-cell stage, 2.93 hpi for the four-cell stage, and 3.04 hpi for the five-cell stage, showing an increasing trend in the time delay with an increasing number of cells. However, the methodology also detected the starting time in hpi before the embryologists for a few instances of the cell cleavage stages. The embryologists (E1, E2, and E3) manually inspected these cases and found that the most of these instances were in the transition phase or capturing active cell division. The embryologists concluded that the methodology’s prediction was correct around the transition phase, which is also subjective and challenging to annotate. The embryologists further pointed out that the primary reason that the methodology reported time delays was the presence of excessive overlapping between cell membranes in the video. Naturally, the overlapping becomes increasingly challenging with the increasing number of cells in the embryo.
7. Discussion
This study aimed to propose a methodology that could help embryologists in clinical settings to annotate the start time of human embryo cell cleavage stages post insemination. Evaluating the quality of embryos involves analyzing the time duration of different cleavage stages, and automating the process of time computation would be useful. Our presented methodology can detect the starting time for cell cleavage stages up to five cells with a delay of 2–3 h post insemination (hpi). When the cell count during cleavage stages was higher than five, the methodology experienced a significant time delay compared to the majority of votes from embryologists. This was because there was excessive overlapping between cell boundaries, which became more noticeable as the number of cells increased. This overlapping led to a lower performance of the methodology, and the embryologists confirmed that cell counting becomes difficult when there is excessive overlapping, resulting in higher disagreement amongst themselves. To address the cell counting issue amidst overlapping, a direction for future work is to train the object detection algorithms specifically on frames of cleavage stages with high overlapping between cells. Among the object detection algorithms tested, YOLO v5 outperformed DETR. YOLO employs non-maximum suppression (NMS) to suppress the bounding boxes with high overlapping areas, which could potentially explain its difficulty in detecting distinct cells with overlapping boundaries. The strategy to redesign NMS with soft-NMS also proved to be ineffective. In future studies, we plan to investigate the methodology using YOLO v5 with adaptive NMS or using Confluence [
50] as an alternative to NMS to detect the start of the cell cleavage stages with cell counts greater than five.
The methodology using DETR performed the best when each of the DETR’s architectural blocks were separately finetuned during training. When it comes to detecting cells, the performance of the DETR was only slightly less accurate than that of the YOLO. However, the results might improve further if we trained on more data. Additionally, our methodology used a parameter called “threshold” in predicting the start of a cell cleavage stage and its value was determined empirically after evaluating a small dataset. Therefore, having access to more data could help in providing a more accurate estimate of the true value of this parameter. During the division of an embryo cell, some of the cell’s cytoplasm content is not captured by the daughter cells and instead becomes extracellular material or fragments. The rate of fragmentation can affect the performance of our methodology because these fragments can be mistaken for cells. Our suggested methodology analyzed raw TLT video frames without any image processing to remove fragments, so we evaluated its performance against the fragmentation present in the datasets. This evaluation is relevant for clinical settings as well. We found that the methodology correctly identified cells and did not confuse them with fragments, despite the fact that both datasets (TransferV and FrozenV) contained TLT videos with a range of low to high fragmentation rates. However, the sizes of the fragments in these datasets was small to medium. To fully validate the methodology for clinical use, there is a need to evaluate the methodology’s performance also for larger-sized fragments. The methodology accurately detected the structure of the morula and blastocyst cell cleavage states. This is also relevant for clinical use. As a future research direction, we recommend to train the methodology to detect the start of substages for morula (start of compaction) and blastocyst (start, full blastocyst, expansion, and hatching).
The TLT video frames capturing the embryo development only provide two-dimensional images. This imaging limitation results in a loss of depth information regarding the three-dimensional embryo cell structure, making it difficult to identify overlapping cells. To overcome this challenge, we suggest exploring other imaging modalities instead of using time-lapse systems. Currently, the methodology can only predict the start time (in hours post insemination (hpi)) of cell cleavage stages and only for TLT videos. Whether the methodology calculates the hpi or reads it from the video frames, the computation of time is dependent on the time-lapse system. Therefore, switching to a different imaging modality would require updating the methodology to ensure that the time computation for hpi remains accurate.
8. Conclusions
The timing between successive cell cleavages serves as a reliable and noninvasive marker for predicting human embryo viability. Automating the tracking of cell divisions can provide embryologists with valuable insights into embryo development and implantation potential. Our proposed methodology efficiently detected the start of consecutive cell cleavage stages in TLT videos up to the five-cell stage, with an average time delay of 2–3 hpi. Excessive overlapping of cell boundaries led to delays and decreased the performance in detecting the start of later cell cleavage stages (cell count greater than five). However, our methodology accurately detected the distinct structures of cell cleavage stages, such as the morula and blastocyst.
Our approach successfully distinguished between cells and small-sized fragments with overlapping boundaries, preventing the misclassification of fragments as cells. The methodology’s pipeline performed best when YOLO v5 was used for detecting cells, morula, and blastocysts in the TLT video frames and with Pytesseract as the OCR library for reading out time digits. The methodology computed annotations for TLT videos in real time, with the overall computation time for annotating a video being approximately one minute.
For future work, we suggest investigating the latest developments in deep-neural-network-based image analysis to improve the cell detection in overlapping regions. This could involve training object detection algorithms on frames with a high degree of cell overlap, exploring alternative imaging modalities for capturing three-dimensional embryo cell structures, and employing advanced deep learning techniques, such as transformers and capsule networks, to enhance the performance of our methodology in detecting the start of cell cleavage stages with cell counts greater than five.