Abstract
Background: Blood is responsible for delivering nutrients to various organs, which store important health information about the human body. Therefore, the diagnosis of blood can indirectly help doctors judge a person’s physical state. Recently, researchers have applied deep learning (DL) to the automatic analysis of blood cells. However, there are still some deficiencies in these models. Methods: To cope with these issues, we propose a novel network for the multi-classification of blood cells, which is called DLBCNet. A new specifical model for blood cells (BCGAN) is designed to generate synthetic images. The pre-trained ResNet50 is implemented as the backbone model, which serves as the feature extractor. The extracted features are fed to the proposed ETRN to improve the multi-classification performance of blood cells. Results: The average accuracy, average sensitivity, average precision, average specificity, and average f1-score of the proposed model are 95.05%, 93.25%, 97.75%, 93.72%, and 95.38%, accordingly. Conclusions: The performance of the proposed model surpasses other state-of-the-art methods in reported classification results.
1. Introduction
The blood flowing in blood vessels is composed of blood cells and plasma. Blood is red because of red blood cells in the blood. Hemoglobin is a special protein that transports oxygen within red blood cells. It is a protein that makes the blood red and consists of globin and heme. Besides red blood cells, there are also white blood cells and platelets. Although they occupy a small share of blood, their functions are very important. These three kinds of blood cells account for 45% of the blood volume, and the remaining 55% of the volume is plasma.
Blood is distributed throughout the body and delivers nutrients to various organs. Naturally, it also stores important health information about the human body. The blood composition will change when there is a problem in our body. Therefore, the diagnosis of blood can indirectly help doctors judge a person’s physical state, which is the routine blood test we often hear of. The routine blood test mainly includes diagnosing red blood cells, white blood cells, and so on. Its significance is to find many early signs of systemic diseases, diagnose whether there is anemia or blood system disease, and reflect the hematopoietic function of bone marrow. Mainstream blood diagnosis is now used to detect white blood cell abnormalities. White blood cell analysis is an essential examination method for pathological blood samples and is an important indicator for detecting and observing diseases. White blood cell recognition is one of the important components of blood testing. By identifying the total number, relative ratio, and morphology of various white blood cells in the blood, we can determine whether there is a disease, the type of disease, and the severity of the disease. So, examining white blood cells is very important to understanding the body’s condition and diagnosing diseases.
With the unprecedented development of deep learning (DL), scholars have recently applied DL to the automatic analysis of blood cells. Over the past decade, DL methods have been put forward for diagnosing blood cells. Tran et al. [1] introduced a hybrid method to segment blood cells. The proposed method was created with pre-trained VGG-16. The end pooling layer of VGG-16 was replaced with semantic segmentation. The overall accuracy of the proposed method could achieve 89.45% accuracy. Habibzadeh et al. [2] put forward a computer-aided diagnosis (CAD) model to automatically classify blood cells. ResNet and Inception were used for feature extractions. Three technologies were proposed to pre-process images: color distortion, image flipping mirroring, and bounding box distortion. This system yielded 99.46% and 99.84% accuracy with ResNet 101 and ResNet V1 152. Tiwari et al. [3] built a novel model to classify blood cells automatically. There were two convolution layers, two pooling layers, and two fully connected layers. The self-built network achieved 78% accuracy for four categories of classification.
Alzubaidi et al. [4] proposed three self-made DL models to classify red blood cells. These three self-made models were composed of parallel and traditional convolution layers. There were some differences among these three models, such as different numbers of traditional and parallel convolution layers, different filters, and so on. The proposed models yielded 99.54% accuracy and 99.54% accuracy with SVM. Yildirim and Çinar [5] used four different four convolution neural networks (CNNs) with two filters to classify blood cells into four categories. Four CNNs were selected to extract features, which were ResNet50, DenseNet201, AlexNet, and GoogleNet. The median and Gaussian filters were used in this paper. DenseNet201 with a Gauss filter achieved 83.44% accuracy. Delgado-Ortet et al. [6] designed a new clinical decision support system to segment red blood cell images and detect malaria. This system included three steps: the segmentation, cropping, and masking of red blood cells and the classification of malaria. For the segmentation and classification, they designed two novel CNN models. One contained 7 fully convolutional layers, and another one was composed of 13 layers. The segmentation method obtained 93.72% accuracy, and the classification method achieved 87.04% specificity.
Jiang et al. [7] designed a DL model to detect blood cells based on the YOLO. They added the spatial and channel attention mechanisms in the YOLO and named this new network the attention-YOLO. The weighted feature vector replaced the original vector. Khouani et al. [8] proposed a DL model to classify blood cells. Firstly, they pre-processed the input to achieve better performance. Then, they tried five different convolution neural networks: Inception V3, VGG16, VGG19, ResNet50, and ResNet101. ResNet50 with the Adam optimizer could obtain the best performance. The proposed deep learning model obtained 95.73% precision and 0.9706 F-score. Patil et al. [9] introduced a hybrid deep learning model to classify white blood cells, which combined the canonical correlation analysis (CCA) and CNN-LSTM to achieve better performance. When Xception was selected as the backbone model, this system could achieve 95.89% accuracy.
H Mohamed et al. [10] put forward a combined model to classify white blood cells. Some pre-trained CNN models were implemented to extract features, and the traditional machine learning models were selected as the classifier. They tested ten pre-trained CNN models and six traditional machine-learning models. Finally, the MobileNet224 with logistic regression achieved 97.03% accuracy. Rahaman et al. [11] compared two models for detecting and counting blood cells, which were the YOLOv5m and YOLOv5s. Finally, the YOLOv5m and YOLOv5s achieved 0.799 precision and 0.797 precision. Sharma et al. [12] classified blood cells into four types based on DenseNet121. The normalization and data augmentation were implemented to improve the classification performance. This proposed model could achieve 98.84% accuracy, 98.85% sensitivity, and 99.61% specificity.
Aliyu et al. [13] introduced an effective model to classify red blood cells. Two phases were included in this model: firstly, the region of interest (ROI) in blood cells was identified, and secondly, AlexNet was selected for classification. The precision, specificity, sensitivity, and accuracy were 90%, 98.82%, 77%, and 95.92%, respectively. Kassim et al. [14] designed a hybrid pipeline to detect red blood cells. U-Net and Faster R-CNN were the vital parts of this hybrid pipeline. The detection accuracy by the proposed model was 97%. Muthumanjula and Bhoopalan [15] built a novel DL network to detect white blood cells. Firstly, the CMYK-moment approach was implemented to identify ROI. Then, CNN was utilized to achieve features. This novel deep learning network yielded 96.41% accuracy.
Shahin et al. [16] put forward a new method (WBCsNet) to identify white blood cells. Several CNN models were utilized to extract features. The SVM was used as the classifier. The proposed WBCsNet achieved 96.1% accuracy. Ekiz et al. [17] selected two models to detect white blood cells. First, CNNs were applied to extract features. Second, the extracted features were used as the input to SVM for classification. The Con-SVM model could achieve 85.96% accuracy. Ammar et al. [18] applied seven different combinations of CNN models with other traditional classifiers to classify blood cells, including KNN, SVM, and AdaboostM1. Finally, the CNN-AdaboostM1 yielded 88% accuracy.
Singh et al. [19] designed a self-made CNN model which included two convolutional layers, two pooling layers, and two fully connected layers. They tested this self-made CNN with different epochs. When the epoch was chosen as 100, this self-made CNN could obtain 86% accuracy. Liang et al. [20] combined CNN models with other networks for the multi-classification of white blood cells. The pre-trained CNN models were chosen to be the feature extractors. Then, recurrent neural networks were implemented as the classifiers. In the experiments, the Xception-LSTM could achieve 90.79% accuracy.
From the above analysis, a sea of DL models could yield certain blood cell diagnosis performances [21,22,23]. However, there are still some deficiencies in these models. Some of them would use handcrafted features [24,25,26,27], but these features could not be the ideal maps for blood cell diagnosis. Meanwhile, DL models could take a lot of time to complete the experiments because of the massive layers and parameters. Furthermore, the overfitting problem is another major concern when these DL models are trained on medical image datasets, which only contain a small number of images. This paper demonstrates a novel DL model (DLBCNet) for the multi-classification of blood cells. We use pre-trained ResNet50 as the backbone to extract ideal features. There are two ways to deal with the overfitting problem in this paper. First, we propose a new model (BCGAN) to generate synthetic images to create a larger dataset. Second, the proposed ETRN not only has a simpler structure but also achieves better performance than common DL models. The main contributions of our work are given as follows:
- The pre-trained ResNet50 is implemented to extract ideal features by comparing it with other CNN models;
- The proposed BCGAN is used to generate synthetic images to alleviate the overfitting problem;
- We propose ETRN to enhance the robustness with the ensemble strategy of combining three individual networks;
- We propose a novel DL model to classify blood cells, which is named DLBCNet.
2. Materials
The dataset is available on this website (https://www.kaggle.com/datasets/paultimothymooney/blood-cells (accessed on 2 January 2023)). This public blood cell dataset comprises 12,500 augmented images of blood cells. There are four different blood cell classes: neutrophil, eosinophil, monocyte, and lymphocyte. Each blood cell class can have approximately 3000 images. The images of these four classes of blood cells are presented in Figure 1.
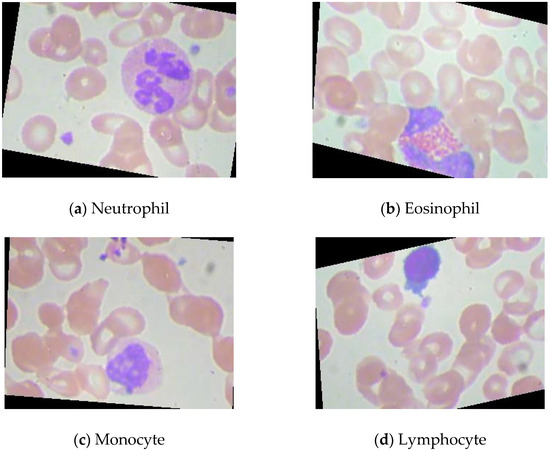
Figure 1.
The images of these four classes of blood cells.
Neutrophils are white blood cells with the highest proportion in peripheral blood, accounting for more than half of white blood cells [28]. They are important components of innate immunity and important effector cells of immune defense. Eosinophils are a kind of white blood cell. Although the number of eosinophils in the blood accounts for less than 5%, they greatly kill bacteria and parasites [29]. Monocytes account for about 3%~8% of the number of white blood cells. They are the largest blood cells in the blood and an important part of the body’s defense system [30]. Lymphocyte is a kind of white blood cell, which is the smallest white blood cell [31]. It is an important cellular component of the immune response function of the body and the main executor of almost all immune functions of the lymphatic system.
3. Methodology
3.1. Feature Learning
Table 1 enumerates the acronyms and provides full explanations. The DL models have achieved remarkable success in various fields, such as natural language processing (NLP), image segmentation, etc. Modern, powerful computing capability makes it possible to have deeper DL networks. These deeper networks often lead to better performance. In recent decades, many epoch-making CNNs have been designed, such as AlexNet [32], ResNet [33], MobileNet [34], and so on.

Table 1.
Acronyms and full explanations.
For image recognition, feature extraction is an important process. Because the volumes of the images are usually too large with excessive information, it is difficult to extract the discrimination rate features. The distribution of features in latent space directly determines the complexity of image classification. With the continuous progress of computer science, CNN models have been the leading solution to the problem of image feature extraction.
It is time-consuming to train CNN models from scratch. Therefore, transfer learning is a feasible method for extracting image features. These pre-trained CNN models are transferred for feature extraction of cell images because they have strong image representation learning ability. ResNet50 is implemented as the backbone model in this paper. The residual connection in ResNet50 is one of the most important inventions in the recent decade of computer science, and can directly connect two non-adjacent layers to complete identity mapping. The framework of the residual connection is given in Figure 2.
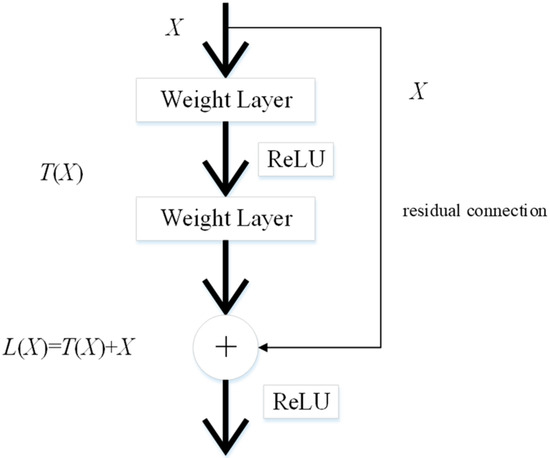
Figure 2.
The structure of the residual connection.
Given as the feature maps from the previous layer, the learned feature is set as . is obtained through the residual connection as follows:
The learned feature with the conversion of the above formula is expressed as follows:
The ResNet50 pre-trained on the ImageNet dataset is modified due to differences in the dataset. The pre-trained ResNet50 is applicable to distinguish 1000 categories of images. Nevertheless, the public blood cell dataset in this paper has only four categories in total: neutrophil, eosinophil, monocyte, and lymphocyte. The modifications to the pre-trained ResNet50 are presented in Figure 3.
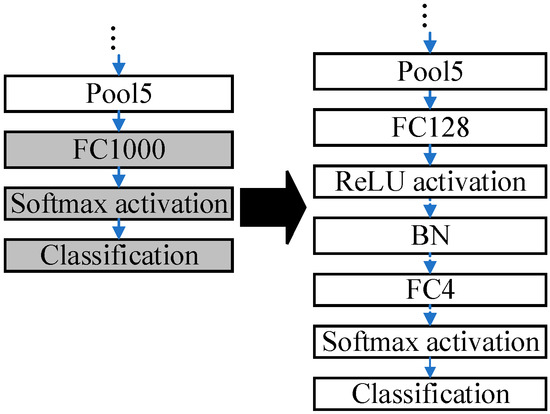
Figure 3.
The modifications to the pre-trained ResNet50.
The last three layers of the pre-trained ResNet50 are removed, and six layers are added, which are ‘FC128’, ‘ReLU’, ‘BN’, ‘FC4’, ‘Softmax’, and ‘Classification’. The parameters in the pre-trained ResNet are frozen except those in the last three layers. Some buffer layers, which are ‘FC128’, ‘ReLU’, and ‘BN’, are inserted between ‘Pool5’ and ‘FC4’ because there are 1000 and 4 output nodes in the ImageNet dataset and the blood cell dataset, accordingly. The buffer layers can smooth the reduction procedures of the dimensions. The modified ResNet50 is fine-tuned by the blood cells dataset.
3.2. Proposed BCGAN
CNN models proved promising when implemented in image recognition and yielded excellent results in big datasets, such as ImageNet [35], CoPhIR [36], and so on. However, the overfitting problem [37] is often encountered when CNN models are applied to small image datasets. The samples of medical datasets are rarely compared with some datasets, such as the ImageNet dataset. It is very time-consuming to create labeled medical datasets.
When researchers employ supervised machine learning models in medical image recognition, the limited labeled dataset can especially restrain the performance. Meanwhile, many studies [38,39,40,41] prove that CNN models can achieve better performance with more data. To deal with these problems, we propose a new generative adversarial network for blood cells (BCGAN) to cope with the limited dataset issue, as shown in Figure 4.
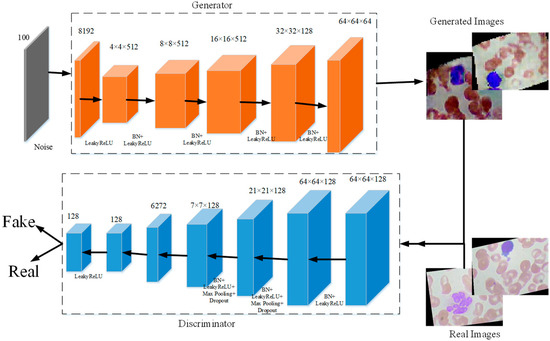
Figure 4.
The proposed BCGAN.
The proposed BCGAN is inspired by generative adversarial networks (GANs) [42]. Two components form the proposed BCGAN, which are the generator and discriminator . The generator obtains the random noise and generates synthetic images. The discriminator is used to identify whether the image is real or fake. The generator and discriminator compete with each other. Generator generates synthetic images similar to the real image as much as possible so that discriminator cannot distinguish the generated images as fake. Discriminator tries to improve the accuracy of identifying the real images and the generated images as much as possible. The proposed BCGAN generates synthetic blood cell images when the discriminator is unable to find the differences between generated images and real images.
Given the data , is denoted as the probability distribution, and the noise is presented as . The loss function is calculated as follows:
where the discriminator tries to maximize from generated data , and the generator is trained to maximize . During the training of the BCGAN, the generator improves its ability to generate more realistic images, and the discriminator enhances the ability to differentiate the real images and generated images. Therefore, the entire training process of BCGAN can be considered as a minimax game between the generator and the discriminator .
In the proposed BCGAN, the convolutional layers are used to extract features. The LeakyReLu is implemented to add nonlinearity. Max pooling is a common strategy to downsample the extracted features. Batch normalization (BN) is chosen to alleviate the gradient disappearance. The overfitting problem can be alleviated by adding the dropout. The BCGAN is specially designed for blood cell images. The pseudocode of the proposed BCGAN is introduced in Algorithm 1. The main contributions of the BCGAN are as follows:
- Five filters are added to increase the ability of the generator to capture more high-level features;
- Additional dropout layers can be helpful in avoiding the overfitting problem;
- The checkboard patterns can be alleviated by the larger kernel size;
- Batch normalization (BN) is inserted into the generator and discriminator to deal with the overfitting problem.
| Algorithm 1 Generative adversarial network for blood cell (BCGAN). |
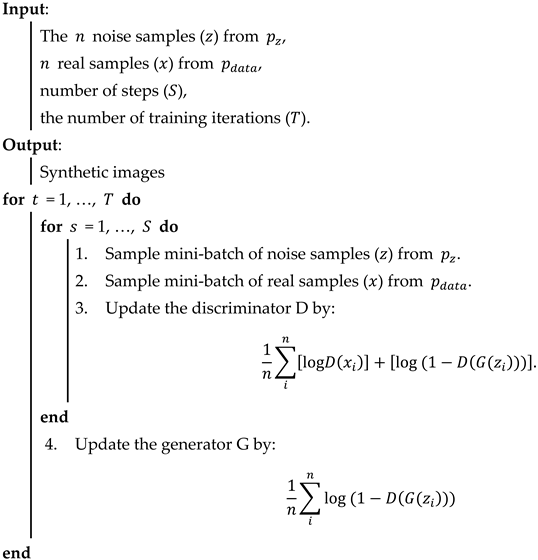 |
We use BCGAN to generate 3000 synthetic images for each class. These synthetic images are mixed with original images to create a new dataset (named mixed-BCGAN dataset). At the same time, we use GANs [42] to generate 3000 synthetic images for each class, which are combined with original images to produce the mixed-GAN dataset.
The comparison of these three datasets is shown in Table 2. The training sets of the mixed-GAN and mixed-BCGAN datasets contain 3000 synthetic images and about 2175 original images for each class. The testing sets of the mixed-GAN and mixed-BCGAN datasets are composed of 933 original images per class. The original dataset’s training set and testing set cover about 2178 and 933 original images per class, respectively.

Table 2.
The comparison of the original dataset with the mixed dataset.
3.3. Proposed ETRN
For the classification of blood cells, three randomized neural networks (RNNs) are implemented to replace the last five layers of the backbone model: extreme learning machine (ELM) [43], random vector functional link (RVFL) [44], and Schmidt neural network (SNN) [45]. These three RNNs merely include three layers: the input layer, hidden layer, and output layer. The training of RNNs can be faster than traditional CNN models benefiting from the simple structure. Compared with the back-propagation neural network, because the weights and bias in RNNs were randomly initialized and fixed in training and the outputs can be calculated by pseudo-inverse, it is unnecessary to update the parameters based on back-propagation, which can shorten the training time. On the other hand, these three RNNs used to replace the last five layers can improve the classification performance.
Ensembles of neural networks are usually recognized to be more robust and accurate compared with individual networks, even though these individual networks can obtain promising results. RNNs are regarded as unstable networks whose performance greatly varies with small perturbations because of the randomized weights and bias. In this situation, we propose a novel network named ETRN to improve classification performance. The structure of the proposed ETRN is given in Figure 5. The pseudocode of the proposed ETRN is shown in Algorithm 2. In the ETRN, three RNNs are trained and then combined with majority voting. The strategy of the ensemble of three RNNs based on majority voting is given below:
where is the image in the dataset, is represented as the ensemble output, and , , and are denoted as the predictions from ELM, RVFL, and SNN, accordingly.
| Algorithm 2 The pseudocode of the proposed ETRN. |
 |
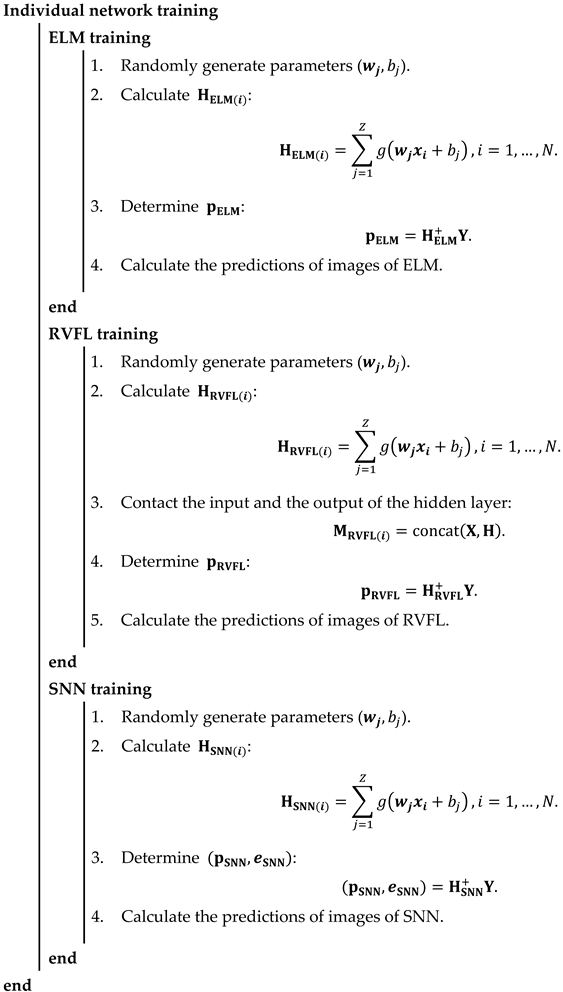 |
 |
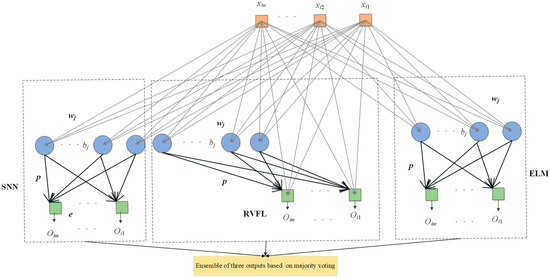
Figure 5.
The structure of ETRN.
The calculations of ELM can be summarized in three steps. Given samples with i-th samples as :
The randomized weights and bias are fixed during the training process, and the outputs of the hidden layer are computed below:
where is the weight between the input and the j-th hidden node, is the bias of the j-th hidden node, is the activation function, and is denoted as the number of hidden nodes.
The output weight is calculated as follows:
where is the pseudo-inverse matrix of and is the ground-truth label matrix of the dataset.
The structure of RVFL has direct connections between the input and output, as shown in Figure 5. Even though the structure is different, the calculation steps are the same. First, calculate the hidden layer output as follows:
The input of the output layer is different because there are direct connections as follows:
where is the input matrix.
The output weight of RVFL is calculated as follows:
The structure of SNN is similar to ELM. The only difference between these two RNNs is that there is an output bias in the SNN. The framework of SNN is presented in Figure 5. The output of the hidden layer in SNN can be computed as follows:
The equation for SNN with output bias is shown below:
3.4. Proposed DLBCNet
We propose a novel DL network to diagnose blood cells (DLBCNet). Collecting a large number of labeled blood cell images to train DL modes is a challenge due to cost and time restrictions. We propose a new specifical model for blood cells (BCGAN) to cope with this challenge. More filters and dropout layers for each layer are added to capture more high-level features. Additional dropout layers and BN are added to avoid the overfitting problem.
Meanwhile, the checkboard patterns can be alleviated by the biggest kernel size. The ResNet50 pre-trained on the ImageNet dataset is implemented as the backbone model in this paper, which is modified and fine-tuned based on blood cells because of the difference between the ImageNet dataset with the blood cell dataset used in this paper. The modified ResNet50 is applied as the feature extractor. The last five layers of the modified ResNet50 are substituted with three RNNs (ELM, RVFL, and SNN). These three RNNs are used for classification. The sample structure and randomized weights of RNNs can reduce training time.
Nevertheless, the RNN is considered an unstable neural network due to some randomized operations. We propose the ETRN by combining three RNNs based on the majority voting to improve the robustness and the generalization performance. The overview of the proposed DLBCNet is demonstrated in Figure 6. The pseudocode of the DLBCNet is illustrated in Algorithm 3.
| Algorithm 3 The pseudocode of the DLBCNet. |
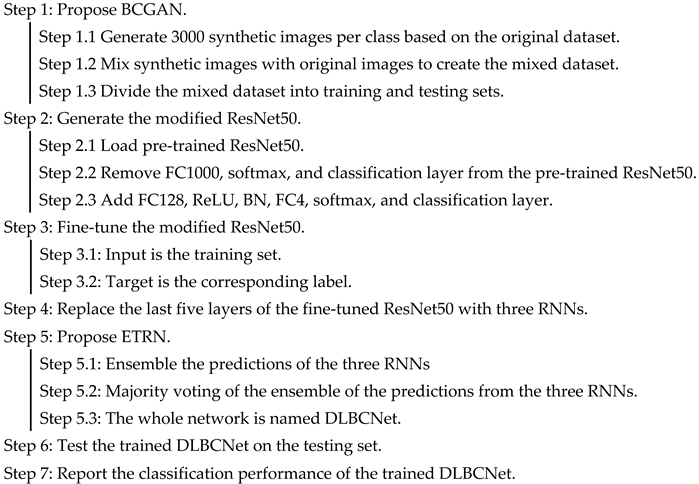 |
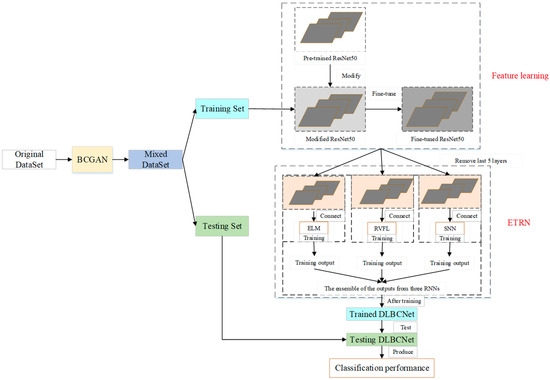
Figure 6.
The overview of the proposed DLBCNet.
3.5. Evaluation
Five multi-classification measurements are applied to evaluate the proposed DLBCNet, which are average accuracy, average sensitivity, average precision, average specificity, and average f1-score for four classes. First, the formulas of accuracy, sensitivity, precision, specificity, and f1-score per class are defined as follows:
where is denoted as the number of classes in this paper. For multi-classification, one class is defined as the positive class. The other three classes are negative classes. The average accuracy, average sensitivity, average precision, average specificity, and average f1-score are calculated below:
The receiver operating characteristic (ROC) curve and the area under the curve (AUC) are used in this paper to evaluate the proposed model.
4. Experiment Settings and Results
4.1. Experiment Settings
The hyper-parameter setting of the proposed DLBCNet is presented in Table 3. The max-epoch is set to 1 to avoid the overfitting problem. The mini-batch size is ten because of the memory size of our device. The initial learning rate is based on experience. The hidden nodes in the hidden layer are set as 400.

Table 3.
The hyper-parameter setting of the proposed DLBCNet.
4.2. The Performance of DLBCNet
Five multi-classification measurements are implemented to evaluate the proposed DLBCNet. Considering the contingency, we carry out five runs. The classification performance of the proposed DLBCNet by five runs is presented in Table 4. The average accuracy, sensitivity, precision, specificity, and f1-score per class by five runs are given in Table 5. The average accuracy, average sensitivity, average precision, average specificity, and average f1-score of the proposed model are 95.05%, 93.25%, 97.75%, 93.72%, and 95.38%, accordingly. All the measurements per class of the proposed DLBCNet are greater than 90%. In particular, our model can achieve promising average accuracy for each class. The ROC curve is presented in Figure 7. The AUC values for eosinophil, lymphocyte, monocyte, and neutrophil are 0.8922, 0.9957, 0.9694, and 0.9091. Generally speaking, it can be concluded that our proposed model is an effective tool for the multi-classification of blood cells.

Table 4.
The performance of the proposed DLBCNet per class (%).

Table 5.
The average multi-classification measurements of the proposed DLBCNet by runs (%).
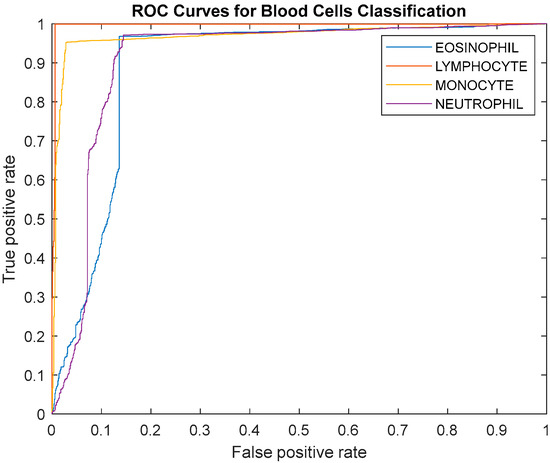
Figure 7.
The ROC curve of the proposed model.
4.3. Comparison of Different Backbone Models
The pre-trained ResNet50 is selected as the backbone model for the proposed DLBCNet. There are numerous famous pre-trained CNN models, such as AlexNet, VGG, ResNet18, and MobileNet. The classification performance of different backbones is demonstrated in Table 6.

Table 6.
The classification performance of different backbones (%).
The proposed DLBCNet with the pre-trained ResNet50 as the backbone model can almost yield the best average accuracy, average sensitivity, average precision, average specificity, and average f1-score compared with other pre-trained models. The residual connection can improve the classification performance. More layers in ResNet50 can extract better features than ResNet18. Therefore, the pre-trained ResNet50 is utilized as the backbone of the proposed DLBCNet.
Using ResNet50 as the backbone model can obtain better results than other backbone models. The reason is that the residual connection in ResNet50 can improve the classification performance. Even though the residual connection is still in ResNet18, deeper layers can extract better features. In this situation, using ResNet50 as the backbone model has better performance than ResNet18.
4.4. Effects of the Proposed BCGAN
The proposed BCGAN is applied to generate synthetic images based on blood cell images to improve the classification performance. We create the mixed-BCGAN dataset based on these synthetic and original images. Meanwhile, the original GANs are compared with the proposed BCGAN to prove its superiority.
The comparison of the classification performance for the mixed and original datasets is demonstrated in Table 7. We test this comparison in five different backbone models to avoid fortuity. These models can yield better classification performance in the mixed-BCGAN dataset than in the mixed-GAN and original datasets. In conclusion, the proposed BCGAN is useful for diagnosing blood cells.

Table 7.
The comparison of the classification performance for the mixed and original datasets (%).
4.5. Effects of RNNs
Three RNNs are implemented as the classifier to replace the last five layers of the backbone model, which are ELM, RVFL, and SNN. The training time of RNNs can be less than traditional CNN models because of the simple structure and fixed randomized parameters. At the same time, RNNs can achieve promising results.
The effects of RNNs are given in Table 8. The classification results using the last five layers are not as good as those using three RNNs. It can be clearly concluded that the three RNNs used to substitute the last five layers can achieve better classification performance. The RNNs can have positive effects on blood cell classification.

Table 8.
The effects of RNNs (%).
4.6. Effects of ETRN
The performance of RNNs can vary with the randomized weights and biases. We propose the ETRN by combining three RNNs to improve classification performance. The effects of the proposed ETRN are shown in Table 9.

Table 9.
The effects of ETRN (%).
The average accuracy per class of ensemble network (DLBCNet) is generally the best, except for eosinophil. The accuracy of eosinophil is only 0.9% less than the best from ResNet50-RVFL. Therefore, the proposed ETRN can improve the multi-classification performance of blood cells.
4.7. Comparison with State-of-the-Art Methods
The proposed DLBCNet is compared to other state-of-the-art methods on the same public dataset, including CNN-AdaboostM1 [18] and the Xception-LSTM [20]. The comparison results of the proposed DLBCNet with other state-of-the-art methods are provided in Table 10.

Table 10.
Comparison with other state-of-the-art methods (%).
Our model can yield the best average accuracy, average sensitivity, average precision, and average f1-score compared with other state-of-the-art methods. The Xception-LSTM achieved the best average specificity of 98.43%, which is 4.7% higher than our model. The comparison results suggest that the proposed DLBCNet is an accurate model for classifying blood cells.
5. Conclusions
The paper put forward a novel network for the classification of blood cells, which is called DLBCNet. We propose a new specifical model for blood cells (BCGAN) to generate synthetic images. The ResNet50 pre-trained on the ImageNet dataset is implemented as the backbone model, which is modified and fine-tuned based on blood cells. The modified ResNet50 is applied as the feature extractor. The extracted features are fed to the proposed ETRN, which combines three RNNs to improve the multi-classification performance of blood cells. The average accuracy, average sensitivity, average precision, average specificity, and average-f1-score of the proposed model are 95.05%, 93.25%, 97.75%, 93.72%, and 95.38%, accordingly.
In future research, we shall apply the proposed model to other public blood cell datasets to prove its generality. Additionally, other recent technology will be implemented in future research, such as MOCO, CLIP, and so on. Moreover, we will try to segment blood cell images.
Author Contributions
Z.Z.: Conceptualization; software; data curation; writing—original draft; writing—review and editing; and visualization. Z.R.: methodology; software; validation; investigation; resources; and writing—review and editing. S.L.: methodology; software; and validation. S.W.: investigation; resources; and writing—review and editing. Y.Z.: methodology; formal analysis; investigation; data curation; writing—original draft; writing—review and editing; supervision; project administration; and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This paper is partially supported by MRC, UK (MC_PC_17171); Royal Society, UK (RP202G0230); BHF, UK (AA/18/3/34220); Hope Foundation for Cancer Research, UK (RM60G0680); GCRF, UK (P202PF11); Sino-UK Industrial Fund, UK (RP202G0289); LIAS, UK (P202ED10, P202RE969); Data Science Enhancement Fund, UK (P202RE237); Fight for Sight, UK (24NN201); Sino-UK Education Fund, UK (OP202006); BBSRC, UK (RM32G0178B8).
Data Availability Statement
The dataset can be downloaded at https://www.kaggle.com/datasets/paultimothymooney/blood-cells (accessed on 2 January 2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tran, T.; Kwon, O.-H.; Kwon, K.-R.; Lee, S.-H.; Kang, K.-W. Blood cell images segmentation using deep learning semantic segmentation. In Proceedings of the 2018 IEEE International Conference on Electronics and Communication Engineering (ICECE), Xi’an, China, 10–12 December 2018; pp. 13–16. [Google Scholar]
- Habibzadeh, M.; Jannesari, M.; Rezaei, Z.; Baharvand, H.; Totonchi, M. Automatic white blood cell classification using pre-trained deep learning models: Resnet and inception. In Proceedings of the Tenth International Conference on Machine Vision (ICMV 2017), Vienna, Austria, 13–15 November 2017; pp. 274–281. [Google Scholar]
- Tiwari, P.; Qian, J.; Li, Q.; Wang, B.; Gupta, D.; Khanna, A.; Rodrigues, J.J.; de Albuquerque, V.H.C. Detection of subtype blood cells using deep learning. Cogn. Syst. Res. 2018, 52, 1036–1044. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Fadhel, M.A.; Al-Shamma, O.; Zhang, J.; Duan, Y. Deep learning models for classification of red blood cells in microscopy images to aid in sickle cell anemia diagnosis. Electronics 2020, 9, 427. [Google Scholar] [CrossRef]
- Yildirim, M.; Çinar, A. Classification of White Blood Cells by Deep Learning Methods for Diagnosing Disease. Rev. D’intelligence Artif. 2019, 33, 335–340. [Google Scholar] [CrossRef]
- Delgado-Ortet, M.; Molina, A.; Alférez, S.; Rodellar, J.; Merino, A. A deep learning approach for segmentation of red blood cell images and malaria detection. Entropy 2020, 22, 657. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, X.; Yan, Z.; Gu, W.; Jiang, J. Improved detection performance in blood cell count by an attention-guided deep learning method. OSA Contin. 2021, 4, 323–333. [Google Scholar] [CrossRef]
- Khouani, A.; El Habib Daho, M.; Mahmoudi, S.A.; Chikh, M.A.; Benzineb, B. Automated recognition of white blood cells using deep learning. Biomed. Eng. Lett. 2020, 10, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Patil, M.; Birajdar, G. White blood cells image classification using deep learning with canonical correlation analysis. Irbm 2021, 42, 378–389. [Google Scholar] [CrossRef]
- Mohamed, E.H.; El-Behaidy, W.H.; Khoriba, G.; Li, J. Improved white blood cells classification based on pre-trained deep learning models. J. Commun. Softw. Syst. 2020, 16, 37–45. [Google Scholar] [CrossRef]
- Rahaman, M.A.; Ali, M.M.; Ahmed, K.; Bui, F.M.; Mahmud, S.H. Performance analysis between YOLOv5s and YOLOv5m model to detect and count blood cells: Deep learning approach. In Proceedings of the 2nd International Conference on Computing Advancements, Dhaka, Bangladesh, 10–12 March 2022; pp. 316–322. [Google Scholar]
- Sharma, S.; Gupta, S.; Gupta, D.; Juneja, S.; Gupta, P.; Dhiman, G.; Kautish, S. Deep learning model for the automatic classification of white blood cells. Comput. Intell. Neurosci. 2022, 2022, 7384131. [Google Scholar] [CrossRef]
- Aliyu, H.A.; Razak, M.A.A.; Sudirman, R.; Ramli, N. A deep learning AlexNet model for classification of red blood cells in sickle cell anemia. Int. J. Artif. Intell. 2020, 9, 221–228. [Google Scholar]
- Kassim, Y.M.; Palaniappan, K.; Yang, F.; Poostchi, M.; Palaniappan, N.; Maude, R.J.; Antani, S.; Jaeger, S. Clustering-based dual deep learning architecture for detecting red blood cells in malaria diagnostic smears. IEEE J. Biomed. Health Inform. 2020, 25, 1735–1746. [Google Scholar] [CrossRef]
- Muthumanjula, M.; Bhoopalan, R. Detection of White Blood Cell Cancer using Deep Learning using Cmyk-Moment Localisation for Information Retrieval. J. IoT Soc. Mob. Anal. Cloud 2022, 4, 54–72. [Google Scholar] [CrossRef]
- Shahin, A.I.; Guo, Y.; Amin, K.M.; Sharawi, A.A. White blood cells identification system based on convolutional deep neural learning networks. Comput. Methods Programs Biomed. 2019, 168, 69–80. [Google Scholar] [CrossRef]
- Ekiz, A.; Kaplan, K.; Ertunç, H.M. Classification of white blood cells using CNN and Con-SVM. In Proceedings of the 2021 29th Signal Processing and Communications Applications Conference (SIU), Istanbul, Turkey, 9–11 June 2021; pp. 1–4. [Google Scholar]
- Ammar, M.; Daho, M.E.H.; Harrar, K.; Laidi, A. Feature extraction using CNN for peripheral blood cells recognition. EAI Endorsed Trans. Scalable Inf. Syst. 2022, 9, e12. [Google Scholar] [CrossRef]
- Singh, I.; Singh, N.P.; Singh, H.; Bawankar, S.; Ngom, A. Blood cell types classification using CNN. In Proceedings of the Bioinformatics and Biomedical Engineering: 8th International Work-Conference, IWBBIO 2020, Granada, Spain, 6–8 May 2020; pp. 727–738. [Google Scholar]
- Liang, G.; Hong, H.; Xie, W.; Zheng, L. Combining convolutional neural network with recursive neural network for blood cell image classification. IEEE Access 2018, 6, 36188–36197. [Google Scholar] [CrossRef]
- Rahali, A.; Akhloufi, M.A. MalBERTv2: Code Aware BERT-Based Model for Malware Identification. Big Data Cogn. Comput. 2023, 7, 60. [Google Scholar] [CrossRef]
- Almotairi, K.H.; Hussein, A.M.; Abualigah, L.; Abujayyab, S.K.; Mahmoud, E.H.; Ghanem, B.O.; Gandomi, A.H. Impact of Artificial Intelligence on COVID-19 Pandemic: A Survey of Image Processing, Tracking of Disease, Prediction of Outcomes, and Computational Medicine. Big Data Cogn. Comput. 2023, 7, 11. [Google Scholar] [CrossRef]
- Chamid, A.A.; Kusumaningrum, R. Graph-Based Semi-Supervised Deep Learning for Indonesian Aspect-Based Sentiment Analysis. Big Data Cogn. Comput. 2022, 7, 5. [Google Scholar] [CrossRef]
- Bogdanchikov, A.; Ayazbayev, D.; Varlamis, I. Classification of Scientific Documents in the Kazakh Language Using Deep Neural Networks and a Fusion of Images and Text. Big Data Cogn. Comput. 2022, 6, 123. [Google Scholar] [CrossRef]
- Bayat, N.; Davey, D.D.; Coathup, M.; Park, J.-H. White Blood Cell Classification Using Multi-Attention Data Augmentation and Regularization. Big Data Cogn. Comput. 2022, 6, 122. [Google Scholar] [CrossRef]
- Moorthy, J.; Gandhi, U.D. A Survey on Medical Image Segmentation Based on Deep Learning Techniques. Big Data Cogn. Comput. 2022, 6, 117. [Google Scholar] [CrossRef]
- Khan, I.U.; Aslam, N.; Anis, F.M.; Mirza, S.; AlOwayed, A.; Aljuaid, R.M.; Bakr, R.M.; Qahtani, N.H.A. Deep Learning-Based Computer-Aided Classification of Amniotic Fluid Using Ultrasound Images from Saudi Arabia. Big Data Cogn. Comput. 2022, 6, 107. [Google Scholar] [CrossRef]
- Nauseef, W.M.; Borregaard, N. Neutrophils at work. Nat. Immunol. 2014, 15, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D.; Foster, P.S. Eosinophils: Changing perspectives in health and disease. Nat. Rev. Immunol. 2013, 13, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef]
- Budd, R.C.; Yeh, W.-C.; Tschopp, J. cFLIP regulation of lymphocyte activation and development. Nat. Rev. Immunol. 2006, 6, 196–204. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Bolettieri, P.; Esuli, A.; Falchi, F.; Lucchese, C.; Perego, R.; Piccioli, T.; Rabitti, F. CoPhIR: A test collection for content-based image retrieval. arXiv 2009, arXiv:0905.4627. [Google Scholar]
- Li, H.; Li, J.; Guan, X.; Liang, B.; Lai, Y.; Luo, X. Research on overfitting of deep learning. In Proceedings of the 2019 15th International Conference on Computational Intelligence and Security (CIS), Macao, China, 13–16 December 2019; pp. 78–81. [Google Scholar]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Ren, Z.; Zhang, Y.; Wang, S. A Hybrid Framework for Lung Cancer Classification. Electronics 2022, 11, 1614. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, Z.; Lv, Z. Medical image synthetic data augmentation using gan. In Proceedings of the 4th International Conference on Computer Science and Application Engineering, New York, NY, USA, 20–22 October 2020; pp. 1–6. [Google Scholar]
- Sedigh, P.; Sadeghian, R.; Masouleh, M.T. Generating synthetic medical images by using GAN to improve CNN performance in skin cancer classification. In Proceedings of the 2019 7th International Conference on Robotics and Mechatronics (ICRoM), Tehran, Iran, 20–21 November 2019; pp. 497–502. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Huang, G.-B.; Zhu, Q.-Y.; Siew, C.-K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Pao, Y.-H.; Park, G.-H.; Sobajic, D.J. Learning and generalization characteristics of the random vector functional-link net. Neurocomputing 1994, 6, 163–180. [Google Scholar] [CrossRef]
- Schmidt, W.F.; Kraaijveld, M.A.; Duin, R.P. Feed forward neural networks with random weights. In Proceedings of the International Conference on Pattern Recognition, The Hague, The Netherlands, 30 August–3 September 1992; pp. 1–4. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).