A Survey on Medical Image Segmentation Based on Deep Learning Techniques
Abstract
1. Introduction
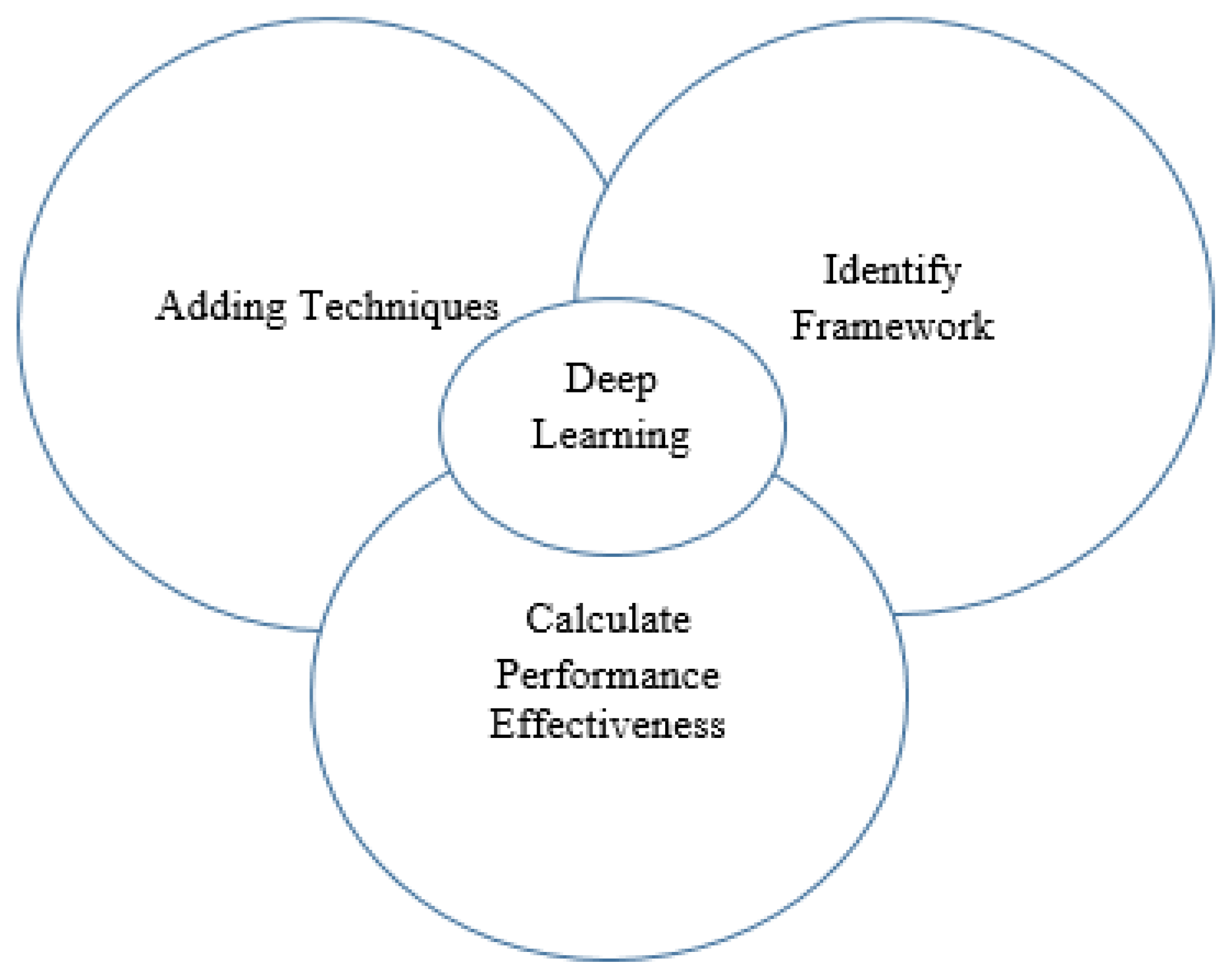
- 1.
- Identify challenges in adding deep learning techniques to the segmentation of medical imaging.
- 2.
- Identify the appropriate framework to remedy or prevent such barriers.
- 3.
- Calculate the overall effectiveness of deep learning approaches with different data sets and frameworks, as shown in Figure 1.
2. Review of Related Work
3. Overview of Deep Learning Techniques
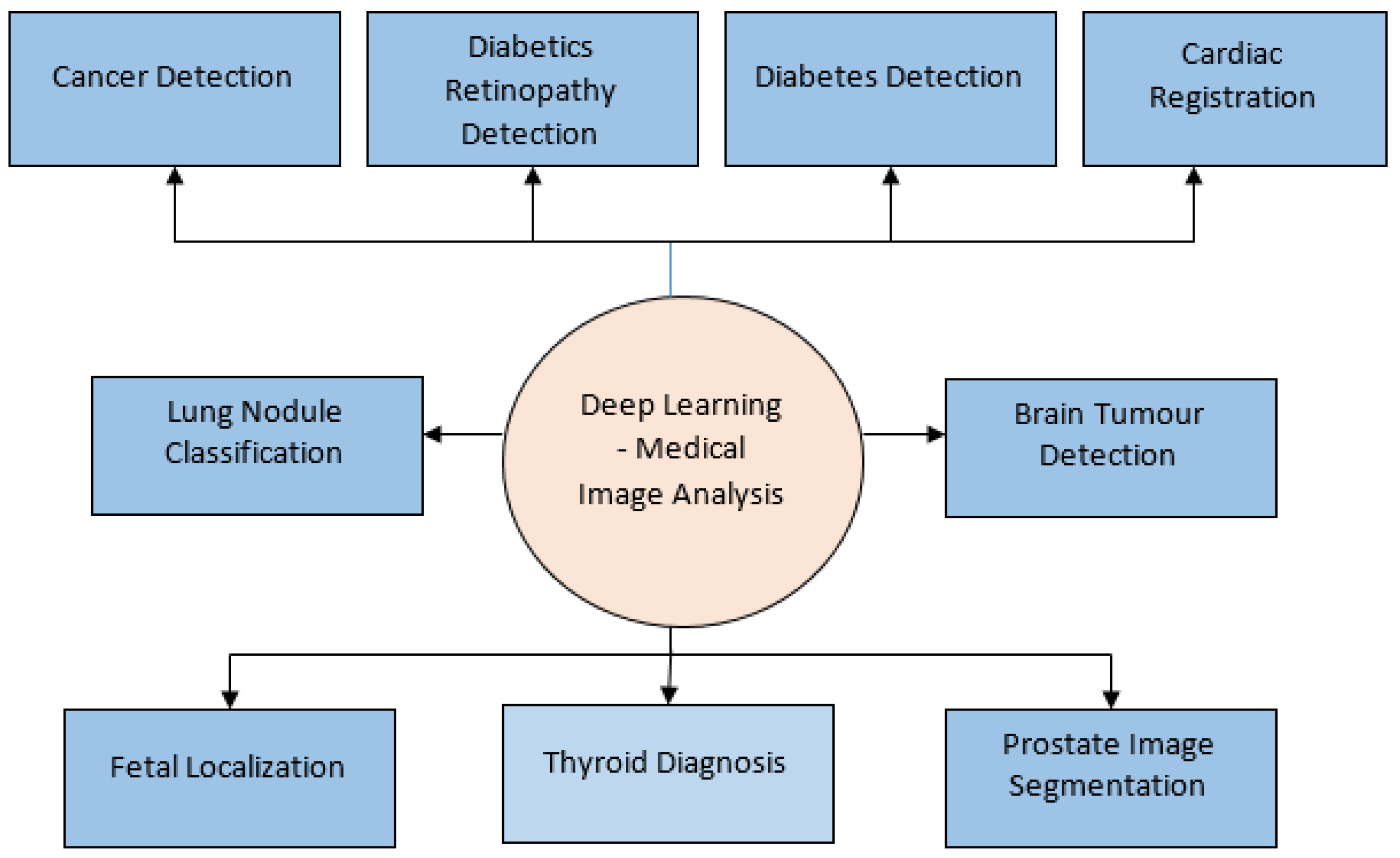
4. Deep Learning Application for Medical Image Analysis
4.1. Segmentation
4.2. Classification
4.3. Detection
4.4. Registration
- 1.
- To implement a continuously proposed methodology, a profound trained model is often used to produce predictors of success between diverse images;
- 2.
- Using deep prediction systems to directly predict transformation parameters. The suggested strategy outperforms state-of-the-art techniques with minimal variables and quick registration durations according to experimental observations.
4.5. Image Enhancement
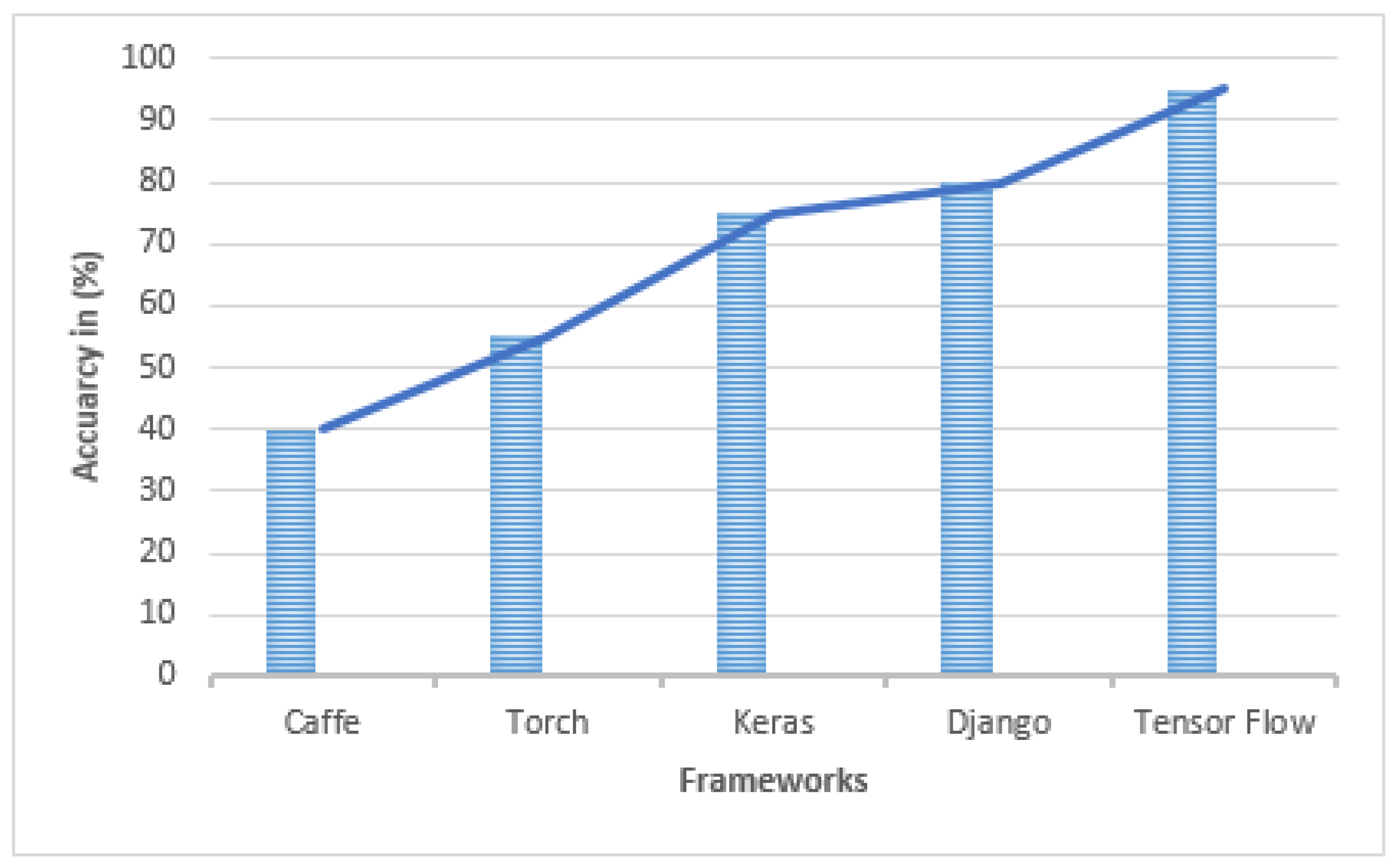
5. Deep Learning Frameworks
- 1.
- Caffe is a deep learning framework for CNN models that is built on MKL, OpenBLAS, and cuBLAS, among other computer packages. Caffe comes with an application of techniques for learning, forecasting, delicate, and so forth. It also includes a number of modeling purposes and procedures for students to use. Caffe’s application servers are simple to install. It also comes with Matlab and Python interfaces that are easy to use. Caffe is simpler to study than other frameworks, which is why many novices prefer it.
- 2.
- TensorFlow is a large-scale platform learning framework that allows machine learning algorithms to be executed through an application. Voice control, data analysis, nanotechnology, knowledge representation, and computational linguistics all seem to be examples of advanced technologies. These are just a few of the applications and off shoots of the making technique to transfer calculations specified using and involving the handling concept onto several device types, including Android and Apple. It also allows processing many devices on a handheld product over several networks.
- 3.
- Torch is capable of supporting the majority of machine learning methods. Multi-layer perceptions, additional frequent approaches, and concepts include SVM classifiers, maximum entropy processes, Markov models, space-time convolutional neural networks, logistic regression, probabilistic classifications, and others. The torch may be integrated into Apple, Android, and FPGA in addition to supporting CPU and GPU.
- 4.
- Keras is a high access soft computing framework built on Programming language. The concept was developed by Francois Chollet, a Microsoft AI-powered developer. Microsoft, Visa, Youtube, Cisco, and Taxi, among several others, are now using Keras. This lecture covers Keras implementations, supervised learning bases, Keras modeling, Keras layering, Keras components, and finally numerous significant implementations.
- 5.
- Django is an elevated Programming language development tool for easily creating secure and functioning websites. Django is a programming environment created by software practitioners that does the hard lifting so you can work on designing your program rather than spinning the wheel. It is freely available, with a thriving community, huge backing, and a range of free model support options.
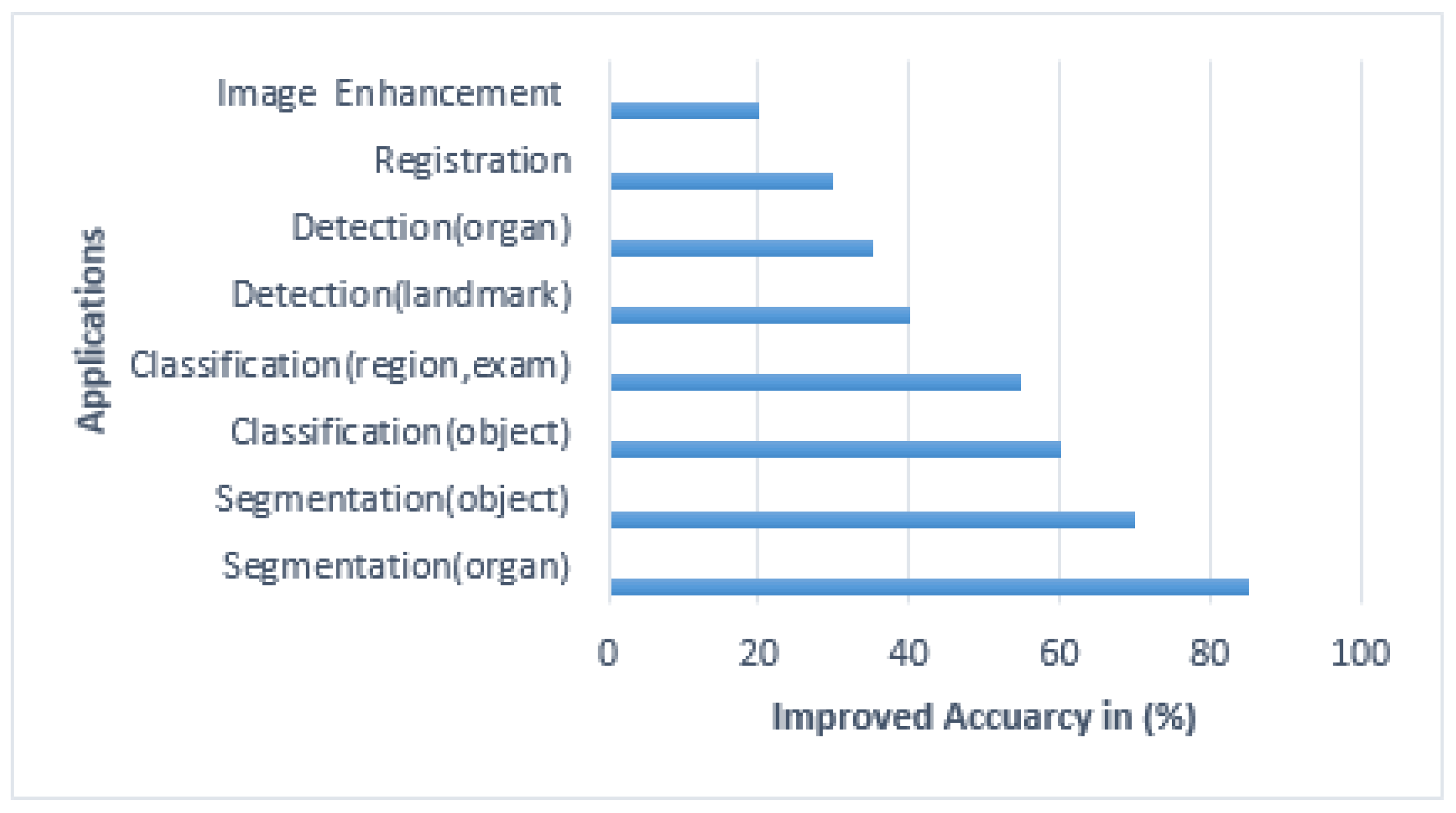
6. Performance Efficiency
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNN | Convolutional Neural Networks |
| AI | Artificial Intelligence |
| DL | Deep Learning |
| RBM | Restricted Boltzmann Machine |
| DNN | Deep Neural Networks |
| RNN | Residual Neural Networks |
| GAN | Generative Adversarial Networks |
| LSTM | Long Short-Term memory |
| DBN | Deep Belief Network |
| DDRL | Direct Deep Reinforcement Learning |
| UST | Therapeutic Ultrasound |
| CT | Computed tomography |
| ANN | Artificial Neural Networks |
| MRI | Magnetic Resonance Imaging |
| MKL | Math Kernel Library |
References
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep learning techniques for medical image segmentation: Achievements and challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, U.; Gandhi, U.D. A survey of big data analytics using machine learning algorithms. Anthol. Big Data Anal. Archit. Appl. 2022, 2022, 655–677. [Google Scholar]
- Luis, F.; Kumar, I.; Vijayakumar, V.; Singh, K.U.; Kumar, A. Identifying the patterns state of the art of deep learning models and computational models their challenges. Multimed. Syst. 2021, 27, 599–613. [Google Scholar]
- Moorthy, U.; Gandhi, U.D. A novel optimal feature selection for medical data classification using CNN based Deep learning. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 3527–3538. [Google Scholar] [CrossRef]
- Chen, S.X.; Ni, Y.Q.; Zhou, L. A deep learning framework for adaptive compressive sensing of high-speed train vibration responses. Struct. Control Health Monit. 2020, 29, E2979. [Google Scholar] [CrossRef]
- Finck, T.; Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. A basic introduction to deep learning for medical image analysis. Sensors 2021, 20, 5097. [Google Scholar]
- Kollem, S.; Reddy, K.R.L.; Rao, D.S. A review of image denoising and segmentation methods based on medical images. Int. J. Mach. Learn. Comput. 2019, 9, 288–295. [Google Scholar] [CrossRef]
- Bir, P.; Balas, V.E. A review on medical image analysis with convolutional neural networks. In Proceedings of the IEEE International Conference on Computing, Power and Communication Technologies, Greater Noida, India, 2–4 October 2020; pp. 870–876. [Google Scholar]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. Journal of medical systems. J. Med. Syst. 2019, 42, 1–13. [Google Scholar]
- Kwekha Rashid, A.S.; Abduljabbar, H.N.; Alhayani, B. Coronavirus disease (COVID-19) cases analysis using machine-learning applications. Appl. Nanosci. 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Anwar Beg, O.; Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Sánchez, C.I. A methodology for extracting retinal blood vessels from fundus images using edge detection. Med. Image Anal. 2017, 42, 60–88. [Google Scholar]
- Lu, L.; Zheng, Y.; Carneiro, G.; Yang, L. A survey on deep learning for big data. Inf. Fusion 2018, 42, 146–157. [Google Scholar]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent residual U-Net for medical image segmentation. Journal of Medical Imaging. J. Med. Imaging 2019, 6, 01006. [Google Scholar] [CrossRef] [PubMed]
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep learning for brain MRI segmentation: State of the art and future directions. J. Digit. Imaging 2017, 30, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Andrade, C.; Teixeira, L.F.; Vasconcelos, M.J.M.; Rosado, L. Data Augmentation Using Adversarial Image-to-Image Translation for the classification techniques of Mobile-Acquired Dermatological Images. J. Imaging 2021, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Decker, Y.; Müller, A.; Németh, E.; Schulz-Schaeffer, W.J.; Fatar, M.; Menger, M.D.; Liu, Y.; Fassbender, K. Analysis of the Convolutional neural netwrok in paraffin-embedded brains. Brain Struct. Funct. 2018, 223, 1001–1015. [Google Scholar] [CrossRef] [PubMed]
- Korte, N.; Nortley, R.; Attwell, D. RNN using cerebral blood flow decrease as an early pathological mechanism in Alzheimer’s disease. Acta Neuropathol. 2020, 40, 793–810. [Google Scholar] [CrossRef] [PubMed]
- Fazlollahi, A.; Calamante, F.; Liang, X.; Bourgeat, P.; Raniga, P.; Dore, V.; Fripp, J.; Ames, D.; Masters, C.L.; Rowe, C.C.; et al. Increased cerebral blood flow with increased amyloid burden in the preclinical phase of alzheimer’s disease. J. Magn. Reson. Imaging 2020, 51, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, B.; Hasan, K.M.; Soares, J.C. Prediction of individual subject’s age across the human lifespan using diffusion tensor imaging: A deep learning approach. Neurobiol. Aging 2013, 75, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Denker, A.; Schmidt, M.; Leuschner, J.; Maass, P. Conditional Invertible Neural Networks for Medical Imaging. J. Imaging 2021, 7, 243. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Van Ginneken, B.; Madabhushi, A. A review of deep learning in medical imaging: Imaging traits, technology trends, case studies with progress highlights, and future promises. Proc. IEEE 2021, 9, 820–838. [Google Scholar] [CrossRef]
- Thong, K.; Ge, C.; Qu, Q.; Gu, I.Y.-H.; Jakola, A. Multi-stream multi-scale deep convolutional networks for Alzheimer’s disease detection using CT images. Neurocomputing 2016, 350, 60–69. [Google Scholar]
- Hu, L.; Koh, J.E.W.; Jahmunah, V.; Pham, T.H.; Oh, S.L.; Ciaccio, E.J.; Acharya, U.R.; Yeong, C.H.; Fabell, M.K.M.; Rahmat, K.; et al. Automated detection of Alzheimer’s disease using bi-directional empirical model decomposition with CT. Pattern Recognit. Lett. 2016, 135, 106–113. [Google Scholar]
- Jiang, F.; Fu, Y.; Gupta, B.B.; Liang, Y.; Rho, S.; Lou, F.; Tian, Z. Deep learning based multi-channel intelligent attack detection for data security. IEEE Trans. Sustain. Comput. 2018, 5, 204–212. [Google Scholar] [CrossRef]
- Hoogi, A.; Beaulieu, C.F.; Cunha, G.M.; Heba, E.; Sirlin, C.B.; Napel, S.; Rubin, D.L. Adaptive local window for level set segmentation of CT and MRI liver lesions. Med. Image Anal. 2017, 37, 46–55. [Google Scholar] [CrossRef]
- Debelee, T.G.; Kebede, S.R.; Schwenker, F.; Shewarega, Z.M. Deep learning in selected cancers’ image analysis—A survey. J. Imaging 2015, 6, 121. [Google Scholar] [CrossRef]
- Razzak, M.I.; Naz, S.; Zaib, A. Deep learning for medical image processing: Overview, challenges and the future. In Classification in BioApps; Springer: Berlin/Heidelberg, Germany, 2018; pp. 323–350. [Google Scholar]
- Inan, M.S.K.; Alam, F.I.; Hasan, R. Deep integrated pipeline of segmentation guided classification of breast cancer from ultrasound images. Biomed. Signal Process. Control 2022, 75, 103553. [Google Scholar] [CrossRef]
- Bai, I.; Hong, S.; Kim, K.; Kim, T. The Design and Implementation of Simulated Threat the cardiac attack for Generator based on MRI images. Korea Inst. Mil. Sci. Technol. 2013, 22, 797–805. [Google Scholar]
- Tsai, A.; Yezzi, W.; Wells, C.; Tempany, D.; Tucker, A.; Fan, W.E.; Grimson, A.; Willsky, w. A shape-based approach to the Cancer registration of medical imagery using level sets. IEEE Trans. Med. Imaging 2003, 22, 137–154. [Google Scholar] [CrossRef]
- Maier, A.; Syben, C.; Lasser, T.; Riess, C. A gentle introduction to deep learning in medical image processing. Z. Med. Phys. 2019, 29, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Sánchez, C.I. A survey on CNN in medical image analysis. Med. Image Anal. 2015, 22, 70–98. [Google Scholar]
- Wang, S.; Eberl, M.; Fulham, Y.; Yin, J.; Chen, D. A likelihood and local constraint level set model for lesion tumor from CT volumes. IEEE Trans. Biomed. Eng. 2015, 60, 2967–2977. [Google Scholar]
- Hu, K.; Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J. The multimodal brain tumor image segmentation benchmark with CNN. IEEE Trans. Med. Image 2016, 34, 1993–2024. [Google Scholar]
- Chen, S.; Ji, C.; Wang, R.; Wu, H. Fully Convolutional Network based on Contrast Information Integration for Dermoscopic Image Segmentation. In Proceedings of the 2020 5th International Conference on Mathematics and Artificial Intelligence, Chengdu, China, 10–13 April 2020; pp. 176–181. [Google Scholar]
- Ozturk, Z.; Li, X.; Huang, H.; Guo, N.; Li, Q. Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans. Radiat. Plasma Med. Sci. 2017, 3, 162–169. [Google Scholar]
- Kumar, S.N.; Lenin Fred, A.; Padmanabhan, P.; Gulyas, B.; Ajay Kumar, H.; Jonisha Miriam, L.R. Deep Learning Algorithms in Medical Image Processing for Cancer Diagnosis: Overview, Challenges and Future. Deep. Learn. Cancer Diagn. 2021, 37–66. [Google Scholar]
- Butt, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Sánchez, C.I. A survey on deep learning and deep neural networks in medical image analysis. Med. Image Anal. 2019, 42, 60–88. [Google Scholar]
- Budd, S.; Robinson, E.C.; Kainz, B. A survey on active learning and human-in-the-loop deep learning for medical image analysis. Med. Imaging Anal. 2021, 71, 102062. [Google Scholar] [CrossRef]
- Paarth Bir, B.; Shevtsov, A.; Dalechina, A.; Krivov, E.; Kostjuchenko, V.; Golanov, A.; Gombolevskiy, V.; Morozov, S.; Belyaev, M. Accelerating 3D Medical Image Segmentation by RNN Target Localization. J. Imaging 2019, 7, 35. [Google Scholar]
- Lekadir, K.; Osuala, R.; Gallin, C.; Lazrak, N.; Kushibar, K.; Tsakou, G.; Martí-Bonmatí, L. FUTURE-AI: Guiding Principles and Consensus Recommendations for Trustworthy Artificial Intelligence in Medical Imaging. arXiv 2021, arXiv:2109.09658. [Google Scholar]
- Xun, S.; Li, D.; Zhu, H.; Chen, M.; Wang, J.; Li, J.; Huang, P. Generative adversarial networks in medical image segmentation: A review. Comput. Biol. Med. 2022, 140, 105063. [Google Scholar] [CrossRef]


| Author Name | Title | Year | Contribution of Work | Advantages | Disadvantage |
|---|---|---|---|---|---|
| Chen et al. [5] | A deep learning framework for adaptive compressive sensing of high-speed train vibration responses | 2020 | Supervised learning, weakly supervised learning. | The design allowsat different levels independently. | Because numerous connections and parameters is enhanced. |
| Finck T. et al. [6] | A basic introduction to deep learning for medical image analysis | 2021 | convolutional neural networks | The networks detect the relevance of characteristics at various levels. | During the process of training is slow. |
| Kollem et al. [7] | A review of image denoising and segmentation methods based on medical images. | 2019 | AI and DL based Framework | Deep learning makes use of enormous volumes of data and time-consuming process is manual | To circumvent the time-consuming process of manual, which necessitates substantial subject expertise. |
| Bir et al. [8] | A review on medical image analysis with convolutional neural networks. | 2020 | DCNN | The dropouts approach used in both analytically cheap and effective. | Failed to address computational constraints. |
| Anwar.et al. [9] | Medical Image Analysis: A Survey of Deep Learning | 2019 | Retinal anatomy segmentation | The minimal storage size provided is often less relevant. | Requires improvement in learning process. |
| Kwekha Rashid et al. [10] | Deep learning Coronavirus disease (COVID-19) cases analysis using | 2021 | Deep Learning-based respiratory disease sensing devices. | The lower labelling detail required during optimisation. | Transformation compute expenses, and memory costs are also included. |
| O. Anwar Bég et al. [11] | A methodology for extracting retinal blood vessels from fundus images using edge detection | 2017 | This method is adaptable for segmenting retinal images and segmenting. | When compared to other techniques, it can provide superior segmentation performance with a shorter runtime. | To get a segmentation picture, a post-processing phase is performed. |
| References | Applications | Modals | Algorithm | Challenges |
|---|---|---|---|---|
| Denker et al. (2021) [21] | Localization | US | CNN | CNN is used with traditional characteristics to locate areas surrounding the organs. |
| Zhou et al. (2021) [22] | Segmentation | CT | ANN | 2-dimensional ANN with 32 × 32 patching, 90images validated. |
| Thong et al. (2016) [23] | Localization | CT | CNN | Combines local patch and slice based CNN. |
| Hu et al. (2016) [24] | Segmentation | CT | CNN | 3D CNN with time-implicit level levels for liver and spleen segmentation. |
| Jiang et al. (2018) [25] | Liver | tumor | CT | F-CNN U-net, coupled fCNN, and dense 3D CRF are all examples of neural networks. |
| Hoogi et al. (2017) [26] | Lesion | CT/MRI | CNN | The possibilities produced by 2D Cnn are utilized to move the segmentation method. |
| References | Applications | Modals | Algorithm | Challenges |
|---|---|---|---|---|
| Debelee et al. (2015) [27] | Localization of prostate | MRI | ANN | Only 55 prostatitis photos were evaluated. |
| Razzak et al. (2018) [28] | Localize the fetal | UST | RBM | Some behaviour is considered that cannot be measured using typical methods. |
| Inan et al. (2022) [29] | Lung cancer detection | MRI | 3D-CNN | Tiny nodules were not detected with great precision. |
| Bai et al. (2013) [30] | Cardiac registration | MRI | DBN | Classification using many atlases Better computing performance is required. |
| Tsai et al. (2018) [31] | Cancer registration | CT | GAN | A small sample with a mild disease. |
| Maier et al. (2019) [32] | Localization | MRI | RBM | Program for 3D dynamic identification Approaches for reducing implementation time really aren’t mentioned. |
| References | Applications | Modals | Algorithm | Challenges |
|---|---|---|---|---|
| Li et al. (2015) [33] | Segmentation | CT | CNN | 3D CNN featuring moment threshold ranges for lung and organ fragmentation. |
| Wang et al. (2015) [34] | Lesion | CT | ANN | Similar strategy is repeated in two-dimensional 15 × 15 update categorization. |
| Hu et al. (2016) [35] | Liver | CT | CNN | Designs of Cnn using maximum entropy variable; excellent SLIVER06 outcomes. |
| Chen et al. (2020) [36] | Lesion | CT | CNN | Two-dimensional Cnn for identification of malignant tumours in follow-up CT using standard CT as feed. |
| Author name | Datasets | Algorithm | Efficiency |
|---|---|---|---|
| Ozturk et al. (2017) [37] | OLS ARIMA | RBM | Accuracy 78.6, Sensitivity 75.09, Specificity 67.4. |
| Kumar, et al. (2021) [38] | Chest X-ray | DBN | Accuracy 87.6, Sensitivity 84.2, Specificity 67.4. |
| Butt et al. (2019) [39] | Electronic medical records | DNN | Accuracy 90.24, Sensitivity 91.02, Specificity 80.55. |
| Budd et al. (2021) [40] | CT scan Images | CNN | Accuracy 98.08, Sensitivity 98.02, Specificity 92.4. |
| Paarth Bir et al. (2019) [41] | MRI scan images | RNN | Accuracy 97.08. |
| Lekadir et al. (2021) [42] | UST scan images | LSTM | Test Accuracy 90.3. |
| Xun, et al. (2022) [43] | CT exams | GAN | Accuracy 89.02, Specificity 91.2. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moorthy, J.; Gandhi, U.D. A Survey on Medical Image Segmentation Based on Deep Learning Techniques. Big Data Cogn. Comput. 2022, 6, 117. https://doi.org/10.3390/bdcc6040117
Moorthy J, Gandhi UD. A Survey on Medical Image Segmentation Based on Deep Learning Techniques. Big Data and Cognitive Computing. 2022; 6(4):117. https://doi.org/10.3390/bdcc6040117
Chicago/Turabian StyleMoorthy, Jayashree, and Usha Devi Gandhi. 2022. "A Survey on Medical Image Segmentation Based on Deep Learning Techniques" Big Data and Cognitive Computing 6, no. 4: 117. https://doi.org/10.3390/bdcc6040117
APA StyleMoorthy, J., & Gandhi, U. D. (2022). A Survey on Medical Image Segmentation Based on Deep Learning Techniques. Big Data and Cognitive Computing, 6(4), 117. https://doi.org/10.3390/bdcc6040117








