Molecular Epidemiology and Surveillance of Human Adenovirus and Rotavirus A Associated Gastroenteritis in Riyadh, Saudi Arabia
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples Collection and Preparation
2.2. Nucleic Acid Extraction and PCR Detection of Enteric Viruses
2.3. Amplicon Purification and Sequencing
2.4. Phylogenetic Analysis
2.5. Statistical Analysis
3. Results
3.1. Clinical Demographics
3.2. Gender- and Age-Based Distribution of HRV and HAdV
3.3. Temporal Distribution of HAdV and HRV
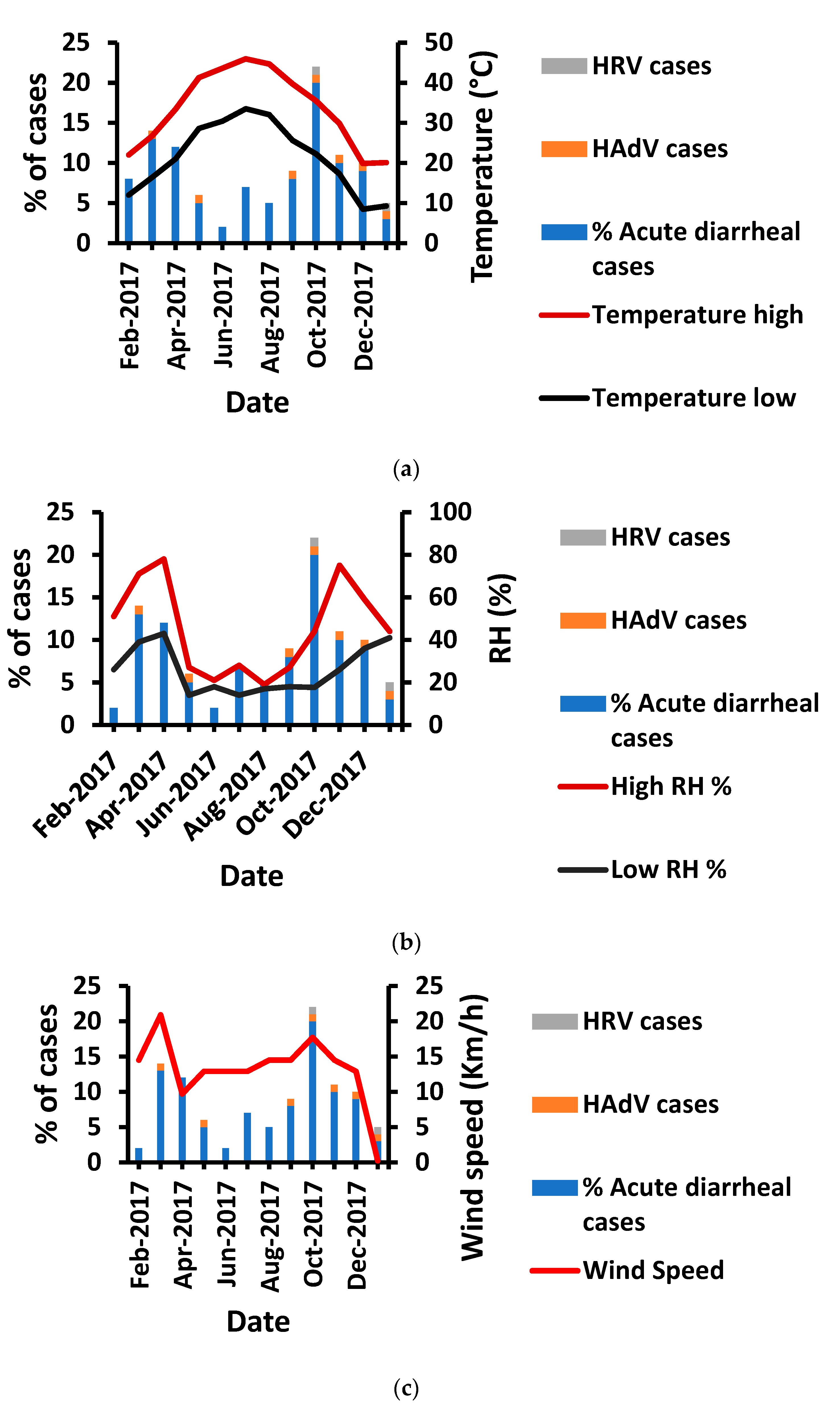
3.4. Temperature Impact on HAdV and HRV Prevalence
3.5. Humidity Impact on HAdV and HRV Prevalence
3.6. Influence of Wind Speed on HRV and HAdV Prevalence
3.7. Prevalence of G2 Lineage of HRV in Patients
3.8. Predominance of HAdV Type 41
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malik, Y.S.; Verma, A.K.; Kumar, N.; Touil, N.; Karthik, K.; Tiwari, R.; Bora, D.P.; Dhama, K.; Ghosh, S.; Hemida, M.G.; et al. Advances in diagnostic approaches for viral etiologies of diarrhea: From the lab to the field. Front. Microbiol. 2019, 10, 1957. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D.; World Health Organization–Coordinated Global Rotavirus Surveillance Network; Agocs, M.; Serhan, F.; de Oliveira, L.; Mwenda, J.M.; Mihigo, R.; et al. Global, regional, and national estimates of rotavirus mortality in children < 5 years of age, 2000–2013. Clin. Infect. Dis. 2016, 62, S96–S105. [Google Scholar] [PubMed]
- Hoffman, S.; Maculloch, B.; Batz, M. Economic burden of major foodborne illnesses acquired in the United States (No. 1476-2016–120935). Econ. Bull. 2015, 140, 4–8. [Google Scholar]
- Lanata, C.F.; Fischer-Walker, C.L.; Olascoaga, A.C.; Torres, C.X.; Aryee, M.J.; Black, R.E.; Child Health Epidemiology Reference Group of the World Health Organization and UNICEF. Global causes of diarrheal disease mortality in children < 5 years of age: A systematic review. PLoS ONE 2013, 8, e72788. [Google Scholar]
- Goel, A.K.; Chawla, S.; Dhingra, A.; Thiyagarajan, V.; Nair, N.P. Rotavirus Diarrhea and Its Determinants among Under-Five Children Admitted in a Tertiary Care Hospital of Southern Haryana, India. Indian J. Pediatr. 2021, 88, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef]
- Nour, I.; Hanif, A.; Ryan, M.; Eifan, S. Insights into Gastrointestinal Virome: Etiology and Public Exposure. Water 2021, 13, 2794. [Google Scholar] [CrossRef]
- O’Ryan, M. The Ever-Changing Landscape of Rotavirus Serotypes. Pediatr. Infect. Dis. J. 2009, 28, S60–S62. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, J.; Amini, R.; Akbari, A.; Amraei, M.; Mahmoudvand, S.; Jalilian, F.A. Prevalence and Seasonal Frequency of Acute Viral Gastroenteritis in Children Less than 5 Years in Ilam, Iran. Prevalence. 2020, 7, 66–74. [Google Scholar]
- Dhingra, A.; Hage, E.; Ganzenmueller, T.; Böttcher, S.; Hofmann, J.; Hamprecht, K.; Obermeier, P.; Rath, B.; Hausmann, F.; Dobner, T. Molecular Evolution of Human Adenovirus (HAdV) Species C. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Lee, B.; Damon, C.F.; Platts-Mills, J.A. Pediatric acute gastroenteritis due to adenovirus 40/41 in low-and middle-income countries. Curr. Opin. Infect. Dis. 2020, 33, 398. [Google Scholar] [CrossRef]
- Lion, T. Adenovirus persistence, reactivation, and clinical management. FEBS Lett. 2019, 593, 3571–3582. [Google Scholar] [CrossRef]
- Tayeb, H.T.; Dela Cruz, D.M.; Al-Qahtani, A.; Al-Ahdal, M.N.; Carter, M.J. Enteric Viruses in Pediatric Diarrhea in Saudi Arabia. J. Med. Virol. 2008, 80, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Meqdam, M.M.; Thwiny, I.R. Prevalence of Group a Rotavirus, Enteric Adenovirus, Norovirus and Astrovirus Infections among Children with Acute Gastroenteritis in Al-Qassim, Saudi Arabia. Pak. J. Med. Sci. 2007, 23, 551. [Google Scholar]
- Akhtar, J.; Qadri, S.M.H.; Myint, S.H. Gastrointestinal Adenovirus Infections in a Tertiary Referral Centre in Saudi Arabia. Eur. J. Clin. Microbiol. Infect. Dis. 1995, 14, 707–710. [Google Scholar] [CrossRef]
- Sdiri-Loulizi, K.; Gharbi-Khélifi, H.; de Rougemont, A.; Chouchane, S.; Sakly, N.; Ambert-Balay, K.; Hassine, M.; Guédiche, M.N.; Aouni, M.; Pothier, P. Acute Infantile Gastroenteritis Associated with Human Enteric Viruses in Tunisia. J. Clin. Microbiol. 2008, 46, 1349–1355. [Google Scholar] [CrossRef]
- Ahmad, S.A.M.; Morsy, A.T.A. Pathogens diarrhea in children, risks and treatment. J. Egypt. Soc. Parasitol. 2022, 52, 278–294. [Google Scholar] [CrossRef]
- Nour, I.; Hanif, A.; Alanazi, I.O.; Al-Ashkar, I.; Alhetheel, A.; Eifan, S. Novel insights of waterborne human rotavirus A in Riyadh (Saudi Arabia) involving G2 predominance and emergence of a thermotolerant sequence. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Dey, R.S.; Ghosh, S.; Chawla-Sarkar, M.; Panchalingam, S.; Nataro, J.P.; Sur, D.; Manna, B.; Ramamurthy, T. Circulation of a Novel Pattern of Infections by Enteric Adenovirus Serotype 41 among Children below 5 Years of Age in Kolkata, India. J. Clin. Microbiol. 2011, 49, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Almalki, S.S. Circulating Rotavirus G and P Strains Post Rotavirus Vaccination in Eastern Mediterranean Region. Saudi Med. J. 2018, 39, 755. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Harastani, H.; Hammadi, M.; Reslan, L.; Ghanem, S.; Hajar, F.; Sabra, A.; Haidar, A.; Inati, A.; Rajab, M. Rotavirus Genotypes and Vaccine Effectiveness from a Sentinel, Hospital-Based, Surveillance Study for Three Consecutive Rotavirus Seasons in Lebanon. PLoS ONE 2016, 11, e0161345. [Google Scholar] [CrossRef] [PubMed]
- Aly, M.; Al Khairy, A.; Al Johani, S.; Balkhy, H. Unusual Rotavirus Genotypes among Children with Acute Diarrhea in Saudi Arabia. BMC Infect. Dis. 2015, 15, 192. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Naghavi, M.; Allen, C.; Barber, R.M.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; Coates, M.M.; Coggeshall, M. Global, Regional, and National Life Expectancy, All-Cause Mortality, and Cause-Specific Mortality for 249 Causes of Death, 1980â 2015: A Systematic Analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef]
- Al Tabbal, A.O.; Al Humedi, S.S. Surveillance of the Most Prevalent Medical Diseases among Pediatric Age Groups and Evaluation of the Control Measures Used At Tabuk Hospitals, Saudi Arabia. Open Access Maced. J. Med. Sci. 2017, 5, 182. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, M.E.; Mathew, S.; Al Thani, A.A.; Ansari, K.A.; Yassine, H.M. Clinical Manifestations Associated with Acute Viral Gastroenteritis Pathogens among Pediatric Patients in Qatar. J. Med. Virol. 2021, 93, 4794–4804. [Google Scholar] [CrossRef]
- Badur, M.; Pidugu, V.K.R.; Kasala, L.; Thiyagarajan, V. Acute Gastroenteritis in Children below 5 Years of Age at Tirupati, Andhra Pradesh, India Post Introduction of Rotavirus Vaccine into National Immunization Programme. Indian J. Pediatr. 2021, 88, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Hegazi, M.A.; Sayed, M.H.; Sindi, H.H.; Bekhit, O.E.; El-Deek, B.S.; Alshoudri, F.M.Y.; Noorelahi, A.K. Is Rotavirus Still a Major Cause for Diarrheal Illness in Hospitalized Pediatric Patients after Rotavirus Vaccine Introduction in the Saudi National Immunization Program? Medicine 2017, 96, e6574. [Google Scholar] [CrossRef] [PubMed]
- Jaff, D.O.; Aziz, T.A.; Smith, N.R. The Incidence of Rotavirus and Adenovirus Infections among Children with Diarrhea in Sulaimani Province, Iraq. J. Biosci. Med. 2015, 4, 124–131. [Google Scholar] [CrossRef]
- Sharif, N.; Parvez, A.K.; Haque, A.; Talukder, A.A.; Ushijima, H.; Dey, S.K. Molecular and Epidemiological Trends of Human Bocavirus and Adenovirus in Children with Acute Gastroenteritis in Bangladesh during 2015 to 2019. J. Med. Virol. 2020, 92, 3194–3201. [Google Scholar] [CrossRef] [PubMed]
- Satter, S.M.; Aliabadi, N.; Gastañaduy, P.A.; Haque, W.; Mamun, A.; Flora, M.S.; Zaman, K.; Rahman, M.; Heffelfinger, J.D.; Luby, S.P. An Update from Hospital-Based Surveillance for Rotavirus Gastroenteritis among Young Children in Bangladesh, July 2012 to June 2017. Vaccine 2018, 36, 7811–7815. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Khurshid, A.; Shaukat, S.; Suleman, R.M.; Sharif, S.; Angez, M.; Malik, S.A.; Ahmed, T.M.; Aamir, U.B.; Naeem, M. Epidemiology and Genetic Diversity of Rotavirus Strains in Children with Acute Gastroenteritis in Lahore, Pakistan. PLoS ONE 2013, 8, e67998. [Google Scholar] [CrossRef]
- Lin, F.J.; Huang, Y.C.; Huang, Y.C.; Huang, L.M.; Liu, C.C.; Chi, H.; Lin, H.C.; Ho, Y.H.; Wu, F.T.; Mu, J.J.; et al. Clinical and epidemiological features in hospitalized young children with acute gastroenteritis in Taiwan: A multicentered surveillance through 2014-2017. J. Formos. Med. Assoc. 2022, 121, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Zeller, M.; Rahman, M.; Heylen, E.; De Coster, S.; De Vos, S.; Arijs, I.; Novo, L.; Verstappen, N.; Van Ranst, M.; Matthijnssens, J. Rotavirus incidence and genotype distribution before and after national rotavirus vaccine introduction in Belgium. Vaccine. 2010, 28, 7507–7513. [Google Scholar] [CrossRef] [PubMed]
- Salami, A.; Fakih, H.; Chakkour, M.; Salloum, L.; Bahmad, H.F.; Ghssein, G. Prevalence, Risk Factors and Seasonal Variations of Different Enteropathogens in Lebanese Hospitalized Children with Acute Gastroenteritis. BMC Pediatr. 2019, 19, 137. [Google Scholar] [CrossRef]
- Okitsu, S.; Khamrin, P.; Hikita, T.; Thongprachum, A.; Pham, N.T.; Hoque, S.A.; Hayakawa, S.; Maneekarn, N.; Ushijima, H. Changing distribution of rotavirus A genotypes circulating in Japanese children with acute gastroenteritis in outpatient clinic, 2014–2020. J. Infect. Public Health 2022, 15, 816–825. [Google Scholar] [CrossRef]
- Al-Ayed, M.S.Z.; Asaad, A.M.; Qureshi, M.A.; Hawan, A.A. Epidemiology of group A rotavirus infection after the introduction of monovalent vaccine in the National Immunization Program of Saudi Arabia. J. Med. Virol. 2017, 89, 429–434. [Google Scholar] [CrossRef]
- Hallowell, B.D.; Parashar, U.D.; Curns, A.; DeGroote, N.P.; Tate, J.E. Trends in the laboratory detection of rotavirus before and after implementation of routine rotavirus vaccination—United States, 2000–2018. Morb. Mortal. Wkly. Rep. 2019, 68, 539. [Google Scholar] [CrossRef] [PubMed]
- Hemming-Harlo, M.; Gylling, A.; Herse, F.; Haavisto, I.; Nuutinen, M.; Pasternack, M.; Kanibir, M.N.; Hartwig, S.; Carias, C. Long-term surveillance of rotavirus vaccination after implementation of a national immunization program in Finland (2008–2018). Vaccine 2022, 40, 3942–3947. [Google Scholar] [CrossRef] [PubMed]
- Kuang, X.; Gong, X.; Zhang, X.; Pan, H.; Teng, Z. Genetic Diversity of Group A Rotavirus in Acute Gastroenteritis Outpatients in Shanghai from 2017 to 2018. BMC Infect. Dis. 2020, 20, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Badur, S.; Öztürk, S.; Pereira, P.; AbdelGhany, M.; Khalaf, M.; Lagoubi, Y.; Ozudogru, O.; Hanif, K.; Saha, D. Systematic Review of the Rotavirus Infection Burden in the WHO-EMRO Region. Hum. Vaccines Immunother. 2019, 15, 2754–2768. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.; Abousekkien, M.; Alkholy, U.M.; Eid, A. Effectiveness and Impact of Rotavirus Vaccines in Saudi Arabia: A Single Hospital-Based Study. Arab J. Gastroenterol. 2017, 18, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Wierzba, T.F.; Abdel-Messih, I.A.; Abu-Elyazeed, R.; Putnam, S.D.; Kamal, K.A.; Rozmajzl, P.; Ahmed, S.F.; Fatah, A.; Zabedy, K.; Shaheen, H.I. Clinic-Based Surveillance for Bacterial-and Rotavirus-Associated Diarrhea in Egyptian Children. Am. J. Trop. Med. Hyg. 2006, 74, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Al-Badani, A.; Al-Areqi, L.; Majily, A.; Al-Sallami, S.; Al-Madhagi, A.; Amood AL-Kamarany, M. Rotavirus Diarrhea among Children in Taiz, Yemen: Prevalence—Risk Factors and Detection of Genotypes. Int. J. Pediatr. 2014, 2014, 928529. [Google Scholar] [CrossRef]
- Biscaro, V.; Piccinelli, G.; Gargiulo, F.; Ianiro, G.; Caruso, A.; Caccuri, F.; De Francesco, M.A. Detection and Molecular Characterization of Enteric Viruses in Children with Acute Gastroenteritis in Northern Italy. Infect. Genet. Evol. 2018, 60, 35–41. [Google Scholar] [CrossRef]
- Nasab, S.D.M.; Zali, F.; Kaghazian, H.; Aghasadeghi, M.R.; Mardani, R.; Gachkar, L.; Vasmehjani, A.A.; Ahmadi, N.; Ghasemzadeh, A. Prevalence of Astrovirus, Adenovirus, and Sapovirus Infections among Iranian Children with Acute Gastroenteritis. Gastroenterol. Hepatol. Bed Bench 2020, 13, S122. [Google Scholar]
- Akello, J.O.; Kamgang, R.; Barbani, M.T.; Suter-Riniker, F.; Leib, S.L.; Ramette, A. Epidemiology of Human Adenoviruses: A 20-Year Retrospective Observational Study in Hospitalized Patients in Bern, Switzerland. Clin. Epidemiol. 2020, 12, 353. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zhang, B.; Xiao, N.; Zhang, F.; Zhao, X.; Liu, Q.; Xie, Z.; Gao, H.; Duan, Z.; Zhong, L. Epidemiology of Human Adenovirus Infection in Children Hospitalized with Lower Respiratory Tract Infections in Hunan, China. J. Med. Virol. 2019, 91, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Arowolo, K.O.; Ayolabi, C.I.; Lapinski, B.; Santos, J.S.; Raboni, S.M. Epidemiology of Enteric Viruses in Children with Gastroenteritis in Ogun State, Nigeria. J. Med. Virol. 2019, 91, 1022–1029. [Google Scholar] [CrossRef] [PubMed]
- D’souza, R.M.; Hall, G.; Becker, N.G. 2008. Climatic factors associated with hospitalizations for rotavirus diarrhoea in children under 5 years of age. Epidemiol. Infect. 2008, 136, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Chong, K.C.; Chan, E.Y.Y.; Lee, T.C.; Kwok, K.L.; Lau, S.Y.F.; Wang, P.; Lam, H.C.Y.; Goggins, W.B.; Mohammad, K.N.; Leung, S.Y. A 21-Year Retrospective Analysis of Environmental Impacts on Paediatric Acute Gastroenteritis in an Affluent Setting. Sci. Total Environ. 2021, 764, 142845. [Google Scholar] [CrossRef]
- Lim, Y.K.; Kweon, O.J.; Kim, H.R.; Kim, T.-H.; Lee, M.-K. Clinical Features, Epidemiology, and Climatic Impact of Genotype-Specific Human Metapneumovirus Infections: Long-Term Surveillance of Hospitalized Patients in South Korea. Clin. Infect. Dis. 2020, 70, 2683–2694. [Google Scholar] [CrossRef] [PubMed]
- Price, R.H.M.; Graham, C.; Ramalingam, S. Association between Viral Seasonality and Meteorological Factors. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Al Musawi, M.; Zainaldeen, H.; Shafi, F.; Anis, S.; DeAntonio, R. Rotavirus Gastroenteritis in Children under 5 Years in the Kingdom of Bahrain: Hospital-Based Surveillance. Clin. Epidemiol. 2013, 5, 269. [Google Scholar]
- Mathew, S.; Al Ansari, K.; Al Thani, A.A.; Zaraket, H.; Yassine, H.M. Epidemiological, Molecular, and Clinical Features of Rotavirus Infections among Pediatrics in Qatar. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1177–1190. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A. The Development of the Qatar Healthcare System: A Review of the Literature. Int. J. Clin. Med. 2015, 6, 177. [Google Scholar] [CrossRef]
- Iaconelli, M.; Valdazo-González, B.; Equestre, M.; Ciccaglione, A.R.; Marcantonio, C.; Della Libera, S.; La Rosa, G. Molecular Characterization of Human Adenoviruses in Urban Wastewaters Using next Generation and Sanger Sequencing. Water Res. 2017, 121, 240–247. [Google Scholar] [CrossRef]

 Refers to HRV sequences (bold italic) belonging to this study.
Refers to HRV sequences (bold italic) belonging to this study.
 Refers to HRV sequences (bold italic) belonging to this study.
Refers to HRV sequences (bold italic) belonging to this study.
 Denotes HAdV sequences (bold italic) belonging to this study.
Denotes HAdV sequences (bold italic) belonging to this study.
 Denotes HAdV sequences (bold italic) belonging to this study.
Denotes HAdV sequences (bold italic) belonging to this study.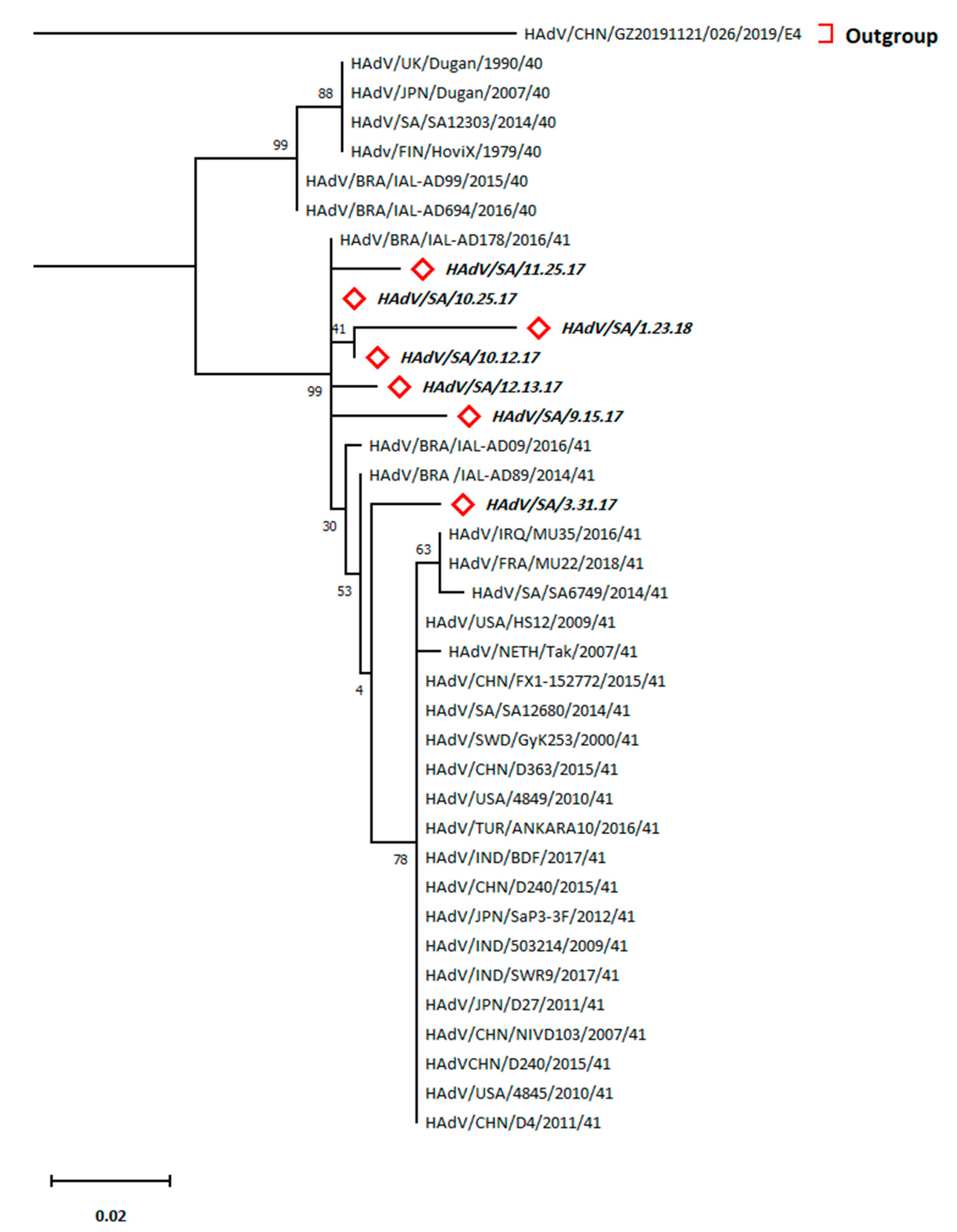
| Age (Years) | HRV Cases | HAdV Cases | ||||
|---|---|---|---|---|---|---|
| Male | Female | Total (%) | Male | Female | Total (%) | |
| <3 | 1/27 (3.7%) | 0/15 (0%) | 1% | 1/27 (3.7%) | 1/15 (6.7%) | 2% |
| 3–5 | 1/9 (11.1%) | 0/10 (0%) | 1% | 0/9 (0%) | 4/10 (40%) | 4% |
| 6–12 | 0/13 (0%) | 0/19 (0%) | 0% | 0/13 (0%) | 0/19 (0%) | 0% |
| 13–18 | 0/1 (0%) | 0/3 (0%) | 0% | 0/1 (0%) | 0/3 (0%) | 0% |
| 19–60 | 0/1 (0%) | 0 | 0% | 1/1 (100%) | 0 | 1% |
| 60+ | 0/1 (0%) | 0/1 (0%) | 0% | 0/1 (0%) | 0/1 (0%) | 0% |
| Total (%) | 2/52 (3.8%) | 0/48 (0%) | 2% | 2/52 (3.8%) | 5/48 (10.4%) | 7% |
| Environmental Factor | Virus and Associated Diseases | R2 | RMSE | Equation ** |
|---|---|---|---|---|
| High temperature (TH) | HAdV | 0.449 | 0.469 | PrevHAdV = 0.34 + 3.3 × 10−2 × TH |
| HRV | 0.002 | 0.534 | PrevHRV = 0.32 − 1.76 × 10−3 × TH | |
| Low temperature (TL) | HAdV | 0.099 | 1.2 | PrevHAdV = 1.52 − 3.46 × 10−2 × TL |
| HRV | 0.134 | 0.497 | PrevHRV = 0.54 − 1.7 × 10−2 × TL | |
| Relative humidity (RH%) | HAdV | 0.140 | 0.830 | PrevHAdV = 1.53 − 1.4 × 10−2 × RH% |
| HRV | 0.216 | 0.473 | PrevHRV = 0.68 − 1.04 × 10−2 × RH% | |
| Wind speed (WS) | HAdV | 0.079 | 1.051 | PrevHAdV = 1.36 − 5.27 × 10−2 × WS |
| HRV | 0.47 | 0.389 | PrevHRV = 0.72 − 6.27 × 10−2 × WS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eifan, S.; Nour, I.; Hanif, A.; Alhetheel, A.; Al-Ashkar, I. Molecular Epidemiology and Surveillance of Human Adenovirus and Rotavirus A Associated Gastroenteritis in Riyadh, Saudi Arabia. Trop. Med. Infect. Dis. 2023, 8, 279. https://doi.org/10.3390/tropicalmed8050279
Eifan S, Nour I, Hanif A, Alhetheel A, Al-Ashkar I. Molecular Epidemiology and Surveillance of Human Adenovirus and Rotavirus A Associated Gastroenteritis in Riyadh, Saudi Arabia. Tropical Medicine and Infectious Disease. 2023; 8(5):279. https://doi.org/10.3390/tropicalmed8050279
Chicago/Turabian StyleEifan, Saleh, Islam Nour, Atif Hanif, Abdulkarim Alhetheel, and Ibrahim Al-Ashkar. 2023. "Molecular Epidemiology and Surveillance of Human Adenovirus and Rotavirus A Associated Gastroenteritis in Riyadh, Saudi Arabia" Tropical Medicine and Infectious Disease 8, no. 5: 279. https://doi.org/10.3390/tropicalmed8050279
APA StyleEifan, S., Nour, I., Hanif, A., Alhetheel, A., & Al-Ashkar, I. (2023). Molecular Epidemiology and Surveillance of Human Adenovirus and Rotavirus A Associated Gastroenteritis in Riyadh, Saudi Arabia. Tropical Medicine and Infectious Disease, 8(5), 279. https://doi.org/10.3390/tropicalmed8050279








