Ecological Niche Modelling Approaches: Challenges and Applications in Vector-Borne Diseases
Abstract
1. Introduction
2. Fundamentals of ENM for VBDs
3. Common Approaches to ENM of VBDs
- The black-box approach integrates across the entire transmission system, assuming that the final distribution of disease cases summarizes all biotic interactions involved in the transmission [44,61]. This approach is useful when transmission dynamics are poorly understood and data are limited [44,66], as happens for many VBDs (e.g., [67,68,69]). However, the black-box oversimplification in such intricate transmission systems may be perilous, as it neglects the ecological requirements of the individual component species [55,66].
- The component-based approach parses the overall transmission cycle into the ecological niches of the individual component species (e.g., pathogens, vectors, hosts) [55]. This approach may allow to distinguish different reasons for presence or absence of disease transmission in an area (i.e., presence/absence of a competent vector and/or susceptible host), but requires potentially lacking in-depth knowledge of the disease system (e.g., the identity and ecologies of relevant species, transmission cycle) [55]. The latter is particularly true for diseases in which the competent vectors might be multiple or poorly known (for instance, see Celone et al. [70], for an approach with the mosquito Haemagogus janthinomys, vector of the Mayaro virus).
4. Relevant Aspects to Consider When Modelling the Niches of VBDs
4.1. Accurate Identification of Occurrence Records
4.2. Global versus Local Occurrence Data
4.3. Importance of Defining M for Background Selection
- (i)
- Being M the geographic extension across which the contrasts should be developed [132], it determines the area within which presences may exist and within which absences are meaningful, in that they represent sites with the broader background landscape actually likely to have been “tested” by the species for suitability, but not occupied [58]. Thus, to minimize the impact of assumptions about absences from areas that are not accessible to the species, the background sample should be chosen to reflect the environmental conditions that one is interested in contrasting against presences [128].
- (ii)
- M has effects on model validation, as areas outside M (where the species cannot occur, owing to restrictions resulting from M but not A) will generally be predicted at lower suitability levels. In consequence, inclusion of these areas (which hold no presence data, but often includes absences that are more distant environmentally from the presences) makes the model look better than it actually is [58].
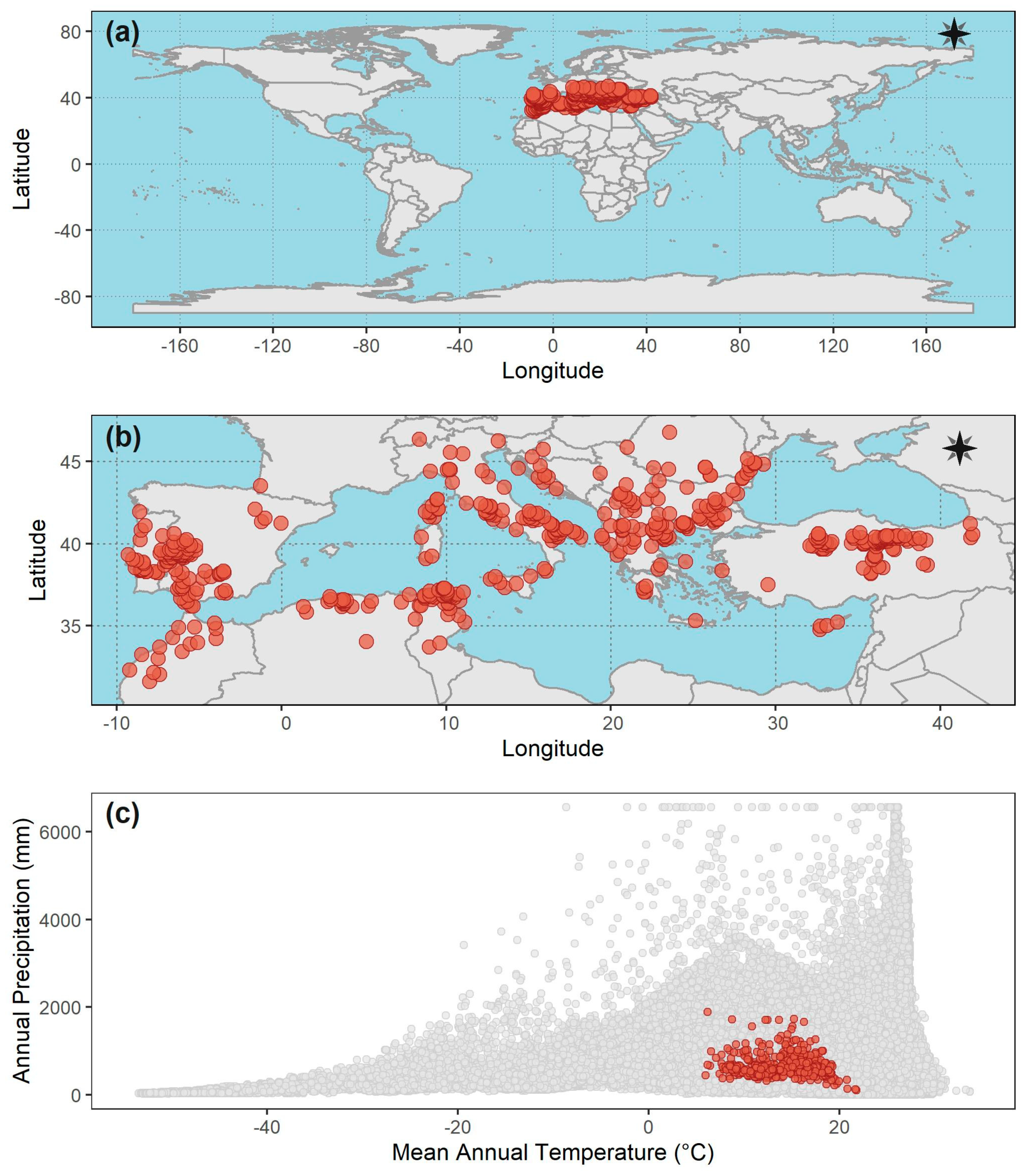
- Geopolitical boundaries: a frequently applied approach is to use administrative boundaries (e.g., municipality, department, province and country) to delimit the calibration area [24,58]. However, this pragmatical use of geopolitical boundaries without an explicit, a priori hypothesis regarding the extent of M presents a major weakness: restricting models based on administrative areas does not account for the biology of the organism [24]. Indeed, most of the cases that applied this approach lacked an explicit assumption about M [67,68,80,83,84,85,86,88,89,91,92,93,94,95,98,99,101,104,106,107,108,109,110,112,116]. Since organisms do not know about geopolitical borders, the perils of this common failure will result in models that are misaligned with the ecology of the organism, resulting in underestimations of the true potential of the disease spread [24] (somehow related to the spatial niche truncation mentioned before).
- Occurrence buffering: an often-used approach consists of determining an optimal buffer area around the occurrences. The radius buffer can be selected in consideration of the dispersal potential of the species [69,81,87,97,101,105,114,117], or after assessing the performance of a series of test models based on buffer with increasing radii (e.g., [100,119]).
- Biogeographical units: these geographical areas are categorized in terms of their biotas [133,134], being defined based on distinct sets of endemic taxa and communities [134]. These areas are usually limited by geographical barriers, altitudinal ranges or a vegetation type [134]. Therefore, the boundaries of the biotic regions within which a species is known to occur may be informative about the barriers that have constrained its distributional potential, resulting in a reasonable hypothesis of the areas that have been available for the species over relevant time periods [58]. To account for potential dispersal beyond these boundaries, an additional buffer can be created around each biogeographical unit occupied [135], or just around the occurrence points within certain distance from the borders of the established biogeographic area [136]. This approach is quite simple, and may prove the most operational [137]. In fact, it has been applied in a number of opportunities to define the accessible area of mosquitoes, kissing bugs and others (i.e., [87,102,117,124,136]).
- Niche-model reconstructions: a simple present-day niche model could be used to estimate the basic dimensions of a species’ distributional potential, and then could be back-projected over the historical conditions that the species has experienced (e.g., Pleistocene Last Glacial Maximum, Last Interglacial) [58,66]. These estimates of past distributional areas can then be combined in a proxy of the long-term dispersal potential, providing a broad initial hypothesis of areas that have been accessible to the species (M) to be used in a second round of model calibration [58,66]. Despite this approach risks some circularity in its implementation, is operational and could be implemented readily [58]. Furthermore, this risk of circularity, owed to the lack of an explicit hypothesis of M in the initial round of modelling [58], could be accounted for by combining the two approaches already described: the biogeographical approach could be used to define M in the initial calibration round, to be later back-projected and used to estimate M in the second round of modelling.
- Full dynamic dispersal models: a more realistic approach should join estimates of the niche with scenarios of dispersal potential through periods of environmental change, considering explicitly the spatially path-dependent nature of effects of environmental change on species’ dispersal reach and consequent distributional potential [58]. Despite its computational challenges, a first akin simulation of this general framework was outlined by Barve et al. [58], and later extended by Machado-Stredel et al. [132]. This intricate approach of defining M is based on a cellular automata simulation where dispersers colonize (or died out) in a cell grid that has suitable conditions drawn from a preliminary estimate of the fundamental ecological niche. These processes are replicated several times to generate an account of accessed cells, to be later summarized in an estimate of M that houses the most frequently accessed cells. This simulation-based method presents a quantitative approach for estimating the accessible area of a species under biologically realistic assumptions, offering the possibility of incorporating relevant climate changes into this estimation of environmental suitability across space and time [132]. As far as we know, this approach has not been applied yet in the modelling of VBDs.
4.4. Sampling Bias and How to Deal with
4.5. Model Complexity and Fine Tuning of the Model Calibration
4.6. Unveiling the Uncertainty Inherent to ENM
- (i)
- the Multivariate Environmental Similarity Surface (MESS) readily incorporated in the Maxent software [169].
- (ii)
5. Main Applications
5.1. Disease Distribution and Risk Mapping
5.2. Filling Gaps in Transmission Cycles
5.3. Assessing the (Potential) Distribution of Invasive Species
5.4. Keeping up with Climate and Global Change: Changes in Time
- (i)
- Effects of niche truncation on model transfers to future climate conditions;
- (ii)
- Effects of model selection procedures on future-climate transfers of ecological niche models;
- (iii)
- Overall variance (uncertainty) in model outcomes.
5.5. Keeping up with Climate and Global Change: Changes in Space
5.6. Aiding to Disentangle Species Complexes
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gubler, D. Vector-borne diseases. Rev. Sci. Tech. 2009, 28, 583–588. [Google Scholar] [CrossRef]
- World Health Organization. Global Vector Control Response 2017–2030; World Health Organization: Geneva, Switzerland, 2017; p. 51. [Google Scholar]
- Roberts, N.L.; Mountjoy-Venning, W.C.; Anjomshoa, M.; Banoub, J.A.M.; Yasin, Y.J. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study. Lancet 2018, 393, E44. [Google Scholar] [CrossRef]
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef]
- Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Stanaway, J.D. The global economic burden of dengue: A systematic analysis. Lancet Infect. Dis. 2016, 16, 935–941. [Google Scholar] [CrossRef]
- Sutherst, R.W. Global Change and Human Vulnerability to Vector-Borne Diseases. Clin. Microbiol. Rev. 2004, 17, 136–173. [Google Scholar] [CrossRef] [PubMed]
- Afshan, K.; Fortes-Lima, C.A.; Artigas, P.; Valero, M.A.; Qayyum, M.; Mas-Coma, S. Impact of climate change and man-made irrigation systems on the transmission risk, long-term trend and seasonality of human and animal fascioliasis in Pakistan. Geospat. Health 2014, 8, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Diuk-Wasser, M.A.; VanAcker, M.C.; Fernandez, M.P. Impact of Land Use Changes and Habitat Fragmentation on the Eco-epidemiology of Tick-Borne Diseases. J. Med. Entomol. 2021, 58, 1546–1564. [Google Scholar] [CrossRef] [PubMed]
- Morand, S.; Lajaunie, C. Outbreaks of Vector-Borne and Zoonotic Diseases Are Associated With Changes in Forest Cover and Oil Palm Expansion at Global Scale. Front. Vet. Sci. 2021, 8, 661063. [Google Scholar] [CrossRef] [PubMed]
- Tamarozzi, F.; Rodari, P.; Salas-Coronas, J.; Bottieau, E.; Salvador, F.; Soriano-Pérez, M.J.; Cabeza-Barrera, M.I.; Van Esbroeck, M.; Treviño, B.; Buonfrate, D.; et al. A large case series of travel-related Mansonella perstans (vector-borne filarial nematode): A TropNet study in Europe. J. Travel Med. 2022, 29, taac048. [Google Scholar] [CrossRef]
- Wilke, A.B.; Benelli, G.; Beier, J.C. Anthropogenic changes and associated impacts on vector-borne diseases. Trends Parasitol. 2021, 37, 1027–1030. [Google Scholar] [CrossRef] [PubMed]
- Caminade, C.; McIntyre, K.M.; Jones, A.E. Impact of recent and future climate change on vector-borne diseases. Ann. N. Y. Acad. Sci. 2019, 1436, 157–173. [Google Scholar] [CrossRef]
- Ogden, N.H.; Lindsay, R. Effects of Climate and Climate Change on Vectors and Vector-Borne Diseases: Ticks Are Different. Trends Parasitol. 2016, 32, 646–656. [Google Scholar] [CrossRef]
- Anyamba, A.; Chretien, J.-P.; Britch, S.C.; Soebiyanto, R.P.; Small, J.L.; Jepsen, R.; Forshey, B.M.; Sanchez, J.L.; Smith, R.D.; Harris, R.; et al. Global Disease Outbreaks Associated with the 2015–2016 El Niño Event. Sci. Rep. 2019, 9, 1930. [Google Scholar] [CrossRef]
- Chretien, J.-P.; Anyamba, A.; Small, J.; Britch, S.; Sanchez, J.L.; Halbach, A.C.; Tucker, C.; Linthicum, K.J. Global Climate Anomalies and Potential Infectious Disease Risks: 2014–2015. PLoS Curr. 2015, 26, 157–173. [Google Scholar] [CrossRef]
- Chambaro, H.M.; Hirose, K.; Sasaki, M.; Libanda, B.; Sinkala, Y.; Fandamu, P.; Muleya, W.; Banda, F.; Chizimu, J.; Squarre, D.; et al. An unusually long Rift valley fever inter-epizootic period in Zambia: Evidence for enzootic virus circulation and risk for disease outbreak. PLoS Negl. Trop. Dis. 2022, 16, e0010420. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [PubMed]
- Mayer, S.V.; Tesh, R.B.; Vasilakis, N. The emergence of arthropod-borne viral diseases: A global prospective on dengue, chikungunya and zika fevers. Acta Trop. 2017, 166, 155–163. [Google Scholar] [CrossRef]
- Messina, J.P.; Brady, O.J.; Golding, N.; Kraemer, M.U.G.; Wint, G.R.W.; Ray, S.E.; Pigott, D.M.; Shearer, F.M.; Johnson, K.; Earl, L.; et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019, 4, 1508–1515. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, M.U.G.; Reiner, R.C., Jr.; Brady, O.J.; Messina, J.P.; Gilbert, M.; Pigott, D.M.; Yi, D.; Johnson, K.; Earl, L.; Marczak, L.B.; et al. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat. Microbiol. 2019, 4, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.J.; Carlson, C.J.; Mordecai, E.A.; Johnson, L.R. Global expansion and redistribution of Aedes-borne virus transmission risk with climate change. PLoS Negl. Trop. Dis. 2019, 13, e0007213. [Google Scholar] [CrossRef] [PubMed]
- Artigas, P.; Reguera-Gomez, M.; Valero, M.A.; Osca, D.; Pacheco, R.D.S.; Rosa-Freitas, M.G.; Silva-Do-Nascimento, T.F.; Paredes-Esquivel, C.; Lucientes, J.; Mas-Coma, S.; et al. Aedes albopictus diversity and relationships in south-western Europe and Brazil by rDNA/mtDNA and phenotypic analyses: ITS-2, a useful marker for spread studies. Parasites Vectors 2021, 14, 333. [Google Scholar] [CrossRef] [PubMed]
- Mas-Coma, S.; Valero, M.A.; Bargues, M.D. Climate change effects on trematodiases, with emphasis on zoonotic fascioliasis and schistosomiasis. Vet. Parasitol. 2009, 163, 264–280. [Google Scholar] [CrossRef]
- Escobar, L.E. Ecological Niche Modeling: An Introduction for Veterinarians and Epidemiologists. Front. Vet. Sci. 2020, 7, 519059. [Google Scholar] [CrossRef] [PubMed]
- Bergquist, R.; Luvall, J.C.; Malone, J.B. The changing risk of vector-borne diseases: Global satellite remote sensing and geospatial surveillance at the forefront. Geospat. Health 2021, 16, 1047. [Google Scholar] [CrossRef]
- Mushegian, A.A.; Neupane, N.; Batz, Z.; Mogi, M.; Tuno, N.; Toma, T.; Miyagi, I.; Ries, L.; Armbruster, P.A. Ecological mechanism of climate-mediated selection in a rapidly evolving invasive species. Ecol. Lett. 2021, 24, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Leimbacher, F. Ecology and control of parasite stages in external environment. Ill. An ecological approach to the control of fascioliasis in France. In Review of Advances in Parasitology; Slusarski, W., Ed.; Polish Scientific Publishers: Warsaw, Poland, 1981; pp. 531–538. [Google Scholar]
- Malone, J.B. Biology-based mapping of vector-borne parasites by Geographic Information Systems and Remote Sensing. Parassitologia 2005, 47, 27–50. [Google Scholar]
- Gillingham, E.L.; Medlock, J.M.; Macintyre, H.; Phalkey, R. Modelling the current and future temperature suitability of the UK for the vector Hyalomma marginatum (Acari: Ixodidae). Ticks Tick-Borne Dis. 2023, 14, 102112. [Google Scholar] [CrossRef]
- Nieto, P.; Malone, J.B.; Bavia, M.E. Ecological niche modeling for visceral leishmaniasis in the state of Bahia, Brazil, using genetic algorithm for rule-set prediction and growing degree day-water budget analysis. Geospat. Health 2006, 1, 115–126. [Google Scholar] [CrossRef]
- Cuervo, P.F.; Fantozzi, M.C.; Di Cataldo, S.; Cringoli, G.; Sierra, R.M.Y.; Rinaldi, L. Analysis of climate and extrinsic incubation of Dirofilaria immitis in southern South America. Geospat. Health 2013, 8, 175–181. [Google Scholar] [CrossRef]
- Cuervo, P.F.; Rinaldi, L.; Cringoli, G. Modeling the extrinsic incubation of Dirofilaria immitis in South America based on monthly and continuous climatic data. Vet. Parasitol. 2015, 209, 70–75. [Google Scholar] [CrossRef]
- Salahi-Moghaddam, A.; Turki, H.; Yeryan, M.; Fuentes, M.V. Spatio-temporal Prediction of the Malaria Transmission Risk in Minab District (Hormozgan Province, Southern Iran). Acta Parasitol. 2022, 67, 1500–1513. [Google Scholar] [CrossRef]
- Ewing, D.A.; Purse, B.V.; Cobbold, C.A.; White, S.M. A novel approach for predicting risk of vector-borne disease establishment in marginal temperate environments under climate change: West Nile virus in the UK. J. R. Soc. Interface 2021, 18, 20210049. [Google Scholar] [CrossRef]
- Paz, S. Climate change impacts on West Nile virus transmission in a global context. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20130561. [Google Scholar] [CrossRef]
- Beesley, N.J.; Caminade, C.; Charlier, J.; Flynn, R.J.; Hodgkinson, J.E.; Martinez-Moreno, A.; Martinez-Valladares, M.; Perez, J.; Rinaldi, L.; Williams, D.J.L. Fasciola and fasciolosis in ruminants in Europe: Identifying research needs. Transbound. Emerg. Dis. 2018, 65, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Beltrame, L.; Dunne, T.; Vineer, H.R.; Walker, J.G.; Morgan, E.R.; Vickerman, P.; McCann, C.M.; Williams, D.J.L.; Wagener, T. A mechanistic hydro-epidemiological model of liver fluke risk. J. R. Soc. Interface 2018, 15, 20180072. [Google Scholar] [CrossRef]
- Estrada-Peña, A.; Ayllón, N.; de la Fuente, J. Impact of Climate Trends on Tick-Borne Pathogen Transmission. Front. Physiol. 2012, 3, 64. [Google Scholar] [CrossRef] [PubMed]
- Tjaden, N.B.; Caminade, C.; Beierkuhnlein, C.; Thomas, S.M. Mosquito-Borne Diseases: Advances in Modelling Climate-Change Impacts. Trends Parasitol. 2018, 34, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Lafferty, K.D. The ecology of climate change and infectious diseases. Ecology 2009, 90, 888–900. [Google Scholar] [CrossRef]
- Anderson, R.P. A framework for using niche models to estimate impacts of climate change on species distributions. Ann. N. Y. Acad. Sci. 2013, 1297, 8–28. [Google Scholar] [CrossRef]
- Peterson, A.T.; Samy, A.M. Geographic potential of disease caused by Ebola and Marburg viruses in Africa. Acta Trop. 2016, 162, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Tjaden, N.B.; Jaeschke, A.; Lühken, R.; Ziegler, U.; Thomas, S.M.; Beierkuhnlein, C. Evaluating the risk for Usutu virus circulation in Europe: Comparison of environmental niche models and epidemiological models. Int. J. Health Geogr. 2018, 17, 35. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.E.; Escobar, L.E.; Zambrana-Torrelio, C. An Ecological Framework for Modeling the Geography of Disease Transmission. Trends Ecol. Evol. 2019, 34, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.T. Biogeography of diseases: A framework for analysis. Sci. Nat. 2008, 95, 483–491. [Google Scholar] [CrossRef]
- Colwell, R.K.; Rangel, T.F. Hutchinson’s duality: The once and future niche. Proc. Natl. Acad. Sci. USA 2009, 106 (Suppl. S2), 19651–19658. [Google Scholar] [CrossRef]
- Soberón, J.; Osorio-Olvera, L.; Peterson, T. Diferencias conceptuales entre modelación de nichos y modelación de áreas de distribución. Rev. Mex. Biodivers. 2017, 88, 437–441. [Google Scholar] [CrossRef]
- Palomar, A.M.; Portillo, A.; Mazuelas, D.; Roncero, L.; Arizaga, J.; Crespo, A.; Gutiérrez, Ó.; Márquez, F.J.; Cuadrado, J.F.; Eiros, J.M.; et al. Molecular analysis of Crimean-Congo hemorrhagic fever virus and Rickettsia in Hyalomma marginatum ticks removed from patients (Spain) and birds (Spain and Morocco), 2009–2015. Ticks Tick-Borne Dis. 2016, 7, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Sadeghieh, T.; Waddell, L.A.; Ng, V.; Hall, A.; Sargeant, J. A scoping review of importation and predictive models related to vector-borne diseases, pathogens, reservoirs, or vectors (1999–2016). PLoS ONE 2020, 15, e0227678. [Google Scholar] [CrossRef]
- Lippi, C.A.; Gaff, H.D.; White, A.L.; Ryan, S.J. Scoping review of distribution models for selected Amblyomma ticks and rickettsial group pathogens. PeerJ 2021, 9, e10596. [Google Scholar] [CrossRef] [PubMed]
- Barker, J.R.; MacIsaac, H.J. Species distribution models applied to mosquitoes: Use, quality assessment, and recommendations for best practice. Ecol. Model. 2022, 472, 110073. [Google Scholar] [CrossRef]
- Kopsco, H.L.; Smith, R.L.; Halsey, S.J. A Scoping Review of Species Distribution Modeling Methods for Tick Vectors. Front. Ecol. Evol. 2022, 10, 893016. [Google Scholar] [CrossRef]
- Molina-Guzmán, L.P.; Gutiérrez-Builes, L.A.; Ríos-Osorio, L.A. Models of spatial analysis for vector-borne diseases studies: A systematic review. Vet. World 2022, 15, 1975–1989. [Google Scholar] [CrossRef]
- Moutinho, S.; Rocha, J.; Gomes, A.; Gomes, B.; Ribeiro, A.I. Spatial Analysis of Mosquito-Borne Diseases in Europe: A Scoping Review. Sustainability 2022, 14, 8975. [Google Scholar] [CrossRef]
- Peterson, A.T.; Soberón, J.; Pearson, R.G.; Anderson, R.P.; Martínez-Meyer, E.; Nakamura, M.; Araújo, M.B. Ecological niches and geographic distributions. In Monographs in Population Biology (Volume 49); Princeton University Press: Princeton, NJ, USA, 2011. [Google Scholar] [CrossRef]
- Soberon, J.; Peterson, A.T. Interpretation of Models of Fundamental Ecological Niches and Species’ Distributional Areas. Biodivers. Inform. 2005, 2, 1–10. [Google Scholar] [CrossRef]
- Soberón, J. Grinnellian and Eltonian niches and geographic distributions of species. Ecol. Lett. 2007, 10, 1115–1123. [Google Scholar] [CrossRef]
- Barve, N.; Barve, V.; Jiménez-Valverde, A.; Lira-Noriega, A.; Maher, S.P.; Townsend Peterson, A.; Soberon, J.; Villalobos, F. The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol. Model. 2011, 222, 1810–1819. [Google Scholar] [CrossRef]
- Owens, H.L.; Campbell, L.P.; Dornak, L.L.; Saupe, E.E.; Barve, N.; Soberón, J.; Ingenloff, K.; Lira-Noriega, A.; Hensz, C.M.; Myers, C.E.; et al. Constraints on interpretation of ecological niche models by limited environmental ranges on calibration areas. Ecol. Model. 2013, 263, 10–18. [Google Scholar] [CrossRef]
- Sillero, N.; Arenas-Castro, S.; Enriquez-Urzelai, U.; Vale, C.G.; Sousa-Guedes, D.; Martínez-Freiría, F.; Real, R.; Barbosa, A. Want to model a species niche? A step-by-step guideline on correlative ecological niche modelling. Ecol. Model. 2021, 456, 109671. [Google Scholar] [CrossRef]
- Peterson, A.T. Ecological niche modelling and understanding the geography of disease transmission. Vet. Ital. 2010, 43, 393–400. [Google Scholar]
- Higgins, S.I.; Larcombe, M.J.; Beeton, N.J.; Conradi, T.; Nottebrock, H. Predictive ability of a process-based versus a correlative species distribution model. Ecol. Evol. 2020, 10, 11043–11054. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Feng, X.; Park, D.S.; Walker, C.; Peterson, A.T.; Merow, C.; Papeş, M. A checklist for maximizing reproducibility of ecological niche models. Nat. Ecol. Evol. 2019, 3, 1382–1395. [Google Scholar] [CrossRef]
- Sillero, N. What does ecological modelling model? A proposed classification of ecological niche models based on their under-lying methods. Ecol. Modell. 2011, 222, 1343–1346. [Google Scholar] [CrossRef]
- Peterson, A.T. Mapping Disease Transmission Risk: Geographic and Ecological Contexts; Johns Hopkins University Press: Baltimore, MD, USA, 2014; p. 210. [Google Scholar]
- Andreo, V.; Rosa, J.; Ramos, K.; Salomón, O.D. Ecological characterization of a cutaneous leishmaniasis outbreak through remotely sensed land cover changes. Geospat. Health 2022, 17, 1033. [Google Scholar] [CrossRef] [PubMed]
- Assefa, A.; Tibebu, A.; Bihon, A.; Dagnachew, A.; Muktar, Y. Ecological niche modeling predicting the potential distribution of African horse sickness virus from 2020 to 2060. Sci. Rep. 2022, 12, 1748. [Google Scholar] [CrossRef]
- Chaves, A.; Dolz, G.; Ibarra-Cerdeña, C.N.; Núñez, G.; Ortiz-Malavasi, E.E.; Bernal-Valle, S.; Gutiérrez-Espeleta, G.A. Presence and potential distribution of malaria-infected New World primates of Costa Rica. Malar. J. 2022, 21, 17. [Google Scholar] [CrossRef]
- Celone, M.; Pecor, D.B.; Potter, A.; Richardson, A.; Dunford, J.; Pollett, S. An ecological niche model to predict the geographic distribution of Haemagogus janthinomys, Dyar, 1921 a yellow fever and Mayaro virus vector, in South America. PLoS Negl. Trop. Dis. 2022, 16, e0010564. [Google Scholar] [CrossRef] [PubMed]
- Escobar, L.E.; Craft, M.E. Advances and Limitations of Disease Biogeography Using Ecological Niche Modeling. Front. Microbiol. 2016, 7, 1174. [Google Scholar] [CrossRef] [PubMed]
- Lozier, J.D.; Aniello, P.; Hickerson, M.J. Predicting the distribution of Sasquatch in western North America: Anything goes with ecological niche modelling. J. Biogeogr. 2009, 36, 1623–1627. [Google Scholar] [CrossRef]
- Harbach, R.E. Culex pipiens: Species Versus Species Complex–Taxonomic History and Perspective. J. Am. Mosq. Control Assoc. 2012, 28 (Suppl. S4), 10–23. [Google Scholar] [CrossRef]
- Tennessen, J.A.; Ingham, V.A.; Toé, K.H.; Guelbéogo, W.M.; Sagnon, N.; Kuzma, R.; Ranson, H.; Neafsey, D.E. A population genomic unveiling of a new cryptic mosquito taxon within the malaria-transmitting Anopheles gambiae complex. Mol. Ecol. 2021, 30, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Walton, C.; Sharpe, R.G.; Pritchard, S.J.; Thelwell, N.J.; Butlin, R.K. Molecular identification of mosquito species. Biol. J. Linn. Soc. 1999, 68, 241–256. [Google Scholar] [CrossRef]
- Mas-Coma, S.; Bargues, M. Populations, hybrids and the systematic concepts of species and subspecies in Chagas disease triatomine vectors inferred from nuclear ribosomal and mitochondrial DNA. Acta Trop. 2009, 110, 112–136. [Google Scholar] [CrossRef] [PubMed]
- Cruz, D.D.; Arellano, E. Molecular data confirm Triatoma pallidipennis Stål, 1872 (Hemiptera: Reduviidae: Triatominae) as a novel cryptic species complex. Acta Trop. 2022, 229, 106382. [Google Scholar] [CrossRef] [PubMed]
- Abad-Franch, F.; Monteiro, F.A.; Pavan, M.G.; Patterson, J.S.; Bargues, M.D.; Zuriaga, M.Á.; Aguilar, M.; Beard, C.B.; Mas-Coma, S.; Miles, M.A. Under pressure: Phenotypic divergence and convergence associated with microhabitat adaptations in Triatominae. Parasites Vectors 2021, 14, 195. [Google Scholar] [CrossRef]
- Gutierrez, M.A.C.; Lopez, R.O.H.; Ramos, A.T.; Vélez, I.D.; Gomez, R.V.; Arrivillaga-Henríquez, J.; Uribe, S. DNA barcoding of Lutzomyia longipalpis species complex (Diptera: Psychodidae), suggests the existence of 8 candidate species. Acta Trop. 2021, 221, 105983. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, M.; Desiato, R.; Albieri, A.; Bellavia, V.; Bertola, M.; Bonilauri, P.; Callegari, E.; Canziani, S.; Lelli, D.; Mosca, A.; et al. Mosquitoes of the Maculipennis complex in Northern Italy. Sci. Rep. 2021, 11, 6421. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, P.F.; Artigas, P.; Mas-Coma, S.; Bargues, M.D. West Nile virus in Spain: Forecasting the geographical distribution of risky areas with an ecological niche modelling approach. Transbound. Emerg. Dis. 2022, 69, e1113–e1129. [Google Scholar] [CrossRef]
- Hernández, C.; Alvarado, M.; Salgado-Roa, F.C.; Ballesteros, N.; Rueda-M, N.; Oliveira, J.; Alevi, K.C.C.; da Rosa, J.A.; Urbano, P.; Salazar, C.; et al. Phylogenetic relationships and evolutionary patterns of the genus Psammolestes Bergroth, 1911 (Hemiptera: Reduviidae: Triatominae). BMC Ecol. Evol. 2022, 22, 30. [Google Scholar] [CrossRef]
- de Beer, C.J.; Dicko, A.H.; Ntshangase, J.; Moyaba, P.; Taioe, M.O.; Mulandane, F.C.; Neves, L.; Mdluli, S.; Guerrini, L.; Bouyer, J.; et al. A distribution model for Glossina brevipalpis and Glossina austeni in Southern Mozambique, Eswatini and South Africa for enhanced area-wide integrated pest management approaches. PLoS Negl. Trop. Dis. 2021, 15, e0009989. [Google Scholar] [CrossRef]
- Fonseca, E.D.S.; Guimarães, R.B.; Prestes-Carneiro, L.E.; Tolezano, J.E.; Rodgers, M.D.S.M.; Avery, R.H.; Malone, J.B. Predicted distribution of sand fly (Diptera: Psychodidae) species involved in the transmission of Leishmaniasis in São Paulo state, Brazil, utilizing maximum entropy ecological niche modeling. Pathog. Glob. Health 2021, 115, 108–120. [Google Scholar] [CrossRef]
- Rodgers, M.D.S.M.; Fonseca, E.; Nieto, P.D.M.; Malone, J.B.; Luvall, J.C.; McCarroll, J.C.; Avery, R.H.; Bavia, M.E.; Guimaraes, R.; Wen, X.; et al. Use of soil moisture active passive satellite data and WorldClim 2.0 data to predict the potential distribution of visceral leishmaniasis and its vector Lutzomyia longipalpis in Sao Paulo and Bahia states, Brazil. Geospat. Health 2022, 17, 1095. [Google Scholar] [CrossRef]
- Fetene, E.; Teka, G.; Dejene, H.; Mandefro, D.; Teshome, T.; Temesgen, D.; Negussie, H.; Mulatu, T.; Jaleta, M.B.; Leta, S. Modeling the spatial distribution of Culicoides species (Diptera: Ceratopogonidae) as vectors of animal diseases in Ethiopia. Sci. Rep. 2022, 12, 12904. [Google Scholar] [CrossRef] [PubMed]
- Flores-López, C.A.; Moo-Llanes, D.A.; Romero-Figueroa, G.; Guevara-Carrizales, A.; López-Ordoñez, T.; Casas-Martínez, M.; Samy, A.M. Potential distributions of the parasite Trypanosoma cruzi and its vector Dipetalogaster maxima highlight areas at risk of Chagas disease transmission in Baja California Sur, Mexico, under climate change. Med. Vet. Entomol. 2022, 36, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Gachoki, S.; Groen, T.; Vrieling, A.; Okal, M.; Skidmore, A.; Masiga, D. Satellite-based modelling of potential tsetse (Glossina pallidipes) breeding and foraging sites using teneral and non-teneral fly occurrence data. Parasites Vectors 2021, 14, 506. [Google Scholar] [CrossRef] [PubMed]
- McBride, S.E.; Lieberthal, B.A.; Buttke, D.E.; Cronk, B.D.; De Urioste-Stone, S.M.; Goodman, L.B.; Guarnieri, L.D.; Rounsville, T.F.; Gardner, A.M. Patterns and Ecological Mechanisms of Tick-Borne Disease Exposure Risk in Acadia National Park, Mount Desert Island, Maine, United States. J. Med. Entomol. 2023, 60, 62–72. [Google Scholar] [CrossRef]
- Moua, Y.; Kotchi, S.; Ludwig, A.; Brazeau, S. Mapping the Habitat Suitability of West Nile Virus Vectors in Southern Quebec and Eastern Ontario, Canada, with Species Distribution Modeling and Satellite Earth Observation Data. Remote Sens. 2021, 13, 1637. [Google Scholar] [CrossRef]
- Nurjanah, S.; Atmowidi, T.; Hadi, U.K.; Solihin, D.D.; Priawandiputra, W.; Santoso, B. Distribution modelling of Aedes aegypti in three dengue-endemic areas in Sumatera, Indonesia. Trop. Biomed. 2022, 39, 373–383. [Google Scholar] [CrossRef]
- Omar, K.; Thabet, H.; TagEldin, R.; Asadu, C.; Chukwuekezie, O.; Ochu, J.; Dogunro, F.; Nwangwu, U.; Onwude, O.; Ezihe, E.; et al. Ecological niche modeling for predicting the potential geographical distribution of Aedes species (Diptera: Culicidae): A case study of Enugu State, Nigeria. Parasite Epidemiol. Control 2021, 15, e00225. [Google Scholar] [CrossRef]
- Springer, A.; Lindau, A.; Probst, J.; Drehmann, M.; Fachet, K.; Thoma, D.; Vineer, H.R.; Noll, M.; Dobler, G.; Mackenstedt, U.; et al. Update and prognosis of Dermacentor distribution in Germany: Nationwide occurrence of Dermacentor reticulatus. Front. Vet. Sci. 2022, 9, 1044597. [Google Scholar] [CrossRef]
- Tagwireyi, P.; Ndebele, M.; Chikurunhe, W. Climate change diminishes the potential habitat of the bont tick (Amblyomma hebraeum): Evidence from Mashonaland Central Province, Zimbabwe. Parasites Vectors 2022, 15, 237. [Google Scholar] [CrossRef]
- Porter, W.T.; Barrand, Z.A.; Wachara, J.; DaVall, K.; Mihaljevic, J.R.; Pearson, T.; Salkeld, D.J.; Nieto, N.C. Predicting the current and future distribution of the western black-legged tick, Ixodes pacificus, across the Western US using citizen science collections. PLoS ONE 2021, 16, e0244754. [Google Scholar] [CrossRef] [PubMed]
- Thameur, B.H.; Soufiène, S.; Ammar, H.H.; Hammami, S. Spatial distribution and habitat selection of culicoides imicola: The potential vector of bluetongue virus in Tunisia. Onderstepoort J. Vet. Res. 2021, 88, 1861. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, P.F.; Flores, F.S.; Venzal, J.M.; Nava, S. Niche divergence among closely related taxa provides insight on evolutionary patterns of ticks. J. Biogeogr. 2021, 48, 2865–2876. [Google Scholar] [CrossRef]
- Rengifo-Correa, L.; González-Salazar, C.; Stephens, C.R. Disentangling the contributions of biotic and abiotic predictors in the niche and the species distribution model of Trypanosoma cruzi, etiological agent of Chagas disease. Acta Trop. 2023, 238, 106757. [Google Scholar] [CrossRef] [PubMed]
- Furlong, M.; Adamu, A.; Hickson, R.I.; Horwood, P.; Golchin, M.; Hoskins, A.; Russell, T. Estimating the Distribution of Japanese Encephalitis Vectors in Australia Using Ecological Niche Modelling. Trop. Med. Infect. Dis. 2022, 7, 393. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, E.D.; Shittu, R.A.; Beierkuhnlein, C.; Thomas, S.M. High Wind Speed Prevents the Establishment of the Disease Vector Mosquito Aedes albopictus in Its Climatic Niche in Europe. Front. Environ. Sci. 2022, 10, 846243. [Google Scholar] [CrossRef]
- Al-Obaidi, M.J.; Ali, H.B. Effect of Climate Change on the Distribution of Zoonotic Cutaneous Leishmaniasis in Iraq. J. Phys. Conf. Ser. 2021, 1818, 12052. [Google Scholar] [CrossRef]
- Campbell, L.; Burkett-Cadena, N.; Miqueli, E.; Unlu, I.; Sloyer, K.; Medina, J.; Vasquez, C.; Petrie, W.; Reeves, L. Potential Distribution of Aedes (Ochlerotatus) scapularis (Diptera: Culicidae): A Vector Mosquito New to the Florida Peninsula. Insects 2021, 12, 213. [Google Scholar] [CrossRef]
- Gorris, M.E.; Bartlow, A.W.; Temple, S.D.; Romero-Alvarez, D.; Shutt, D.P.; Fair, J.M.; Kaufeld, K.A.; Del Valle, S.Y.; Manore, C.A. Updated distribution maps of predominant Culex mosquitoes across the Americas. Parasites Vectors 2021, 14, 547. [Google Scholar] [CrossRef]
- Holeva-Eklund, W.M.; Young, S.J.; Will, J.; Busser, N.; Townsend, J.; Hepp, C.M. Species distribution modeling of Aedes aegypti in Maricopa County, Arizona from 2014 to 2020. Front. Environ. Sci. 2022, 10, 2107. [Google Scholar] [CrossRef]
- Hussain, S.S.A.; Dhiman, R.C. Distribution Expansion of Dengue Vectors and Climate Change in India. Geohealth 2022, 6, e2021GH000477. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, C.B.; Guzmán-Rodríguez, L.; Moreno, M.; Castro, M.D.M.; Valderrama-Ardila, C.; Alexander, N. Integration of phlebotomine ecological niche modelling, and mapping of cutaneous leishmaniasis surveillance data, to identify areas at risk of under-estimation. Acta Trop. 2021, 224, 106122. [Google Scholar] [CrossRef]
- Rhodes, C.G.; Loaiza, J.R.; Romero, L.M.; Alvarado, J.M.G.; Delgado, G.; Salas, O.R.; Rojas, M.R.; Aguilar-Avendaño, C.; Maynes, E.; Cordero, J.A.V.; et al. Anopheles albimanus (Diptera: Culicidae) Ensemble Distribution Modeling: Applications for Malaria Elimination. Insects 2022, 13, 221. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, L.; Ayala, S.; Reyes, C.; González, C.R. Modeling the Potential Distribution of the Malaria Vector Anopheles (Ano.) pseudopunctipennis Theobald (Diptera: Culicidae) in Arid Regions of Northern Chile. Front. Public Health 2021, 9, 611152. [Google Scholar] [CrossRef]
- Sánchez-Fernández, D.; Lobo, J.M.; Hernández-Manrique, O.L. Species distribution models that do not incorporate global data misrepresent potential distributions: A case study using Iberian diving beetles. Divers. Distrib. 2011, 17, 163–171. [Google Scholar] [CrossRef]
- Raes, N. Partial versus Full Species Distribution Models. Nat. Conserv. 2012, 10, 127–138. [Google Scholar] [CrossRef]
- Chevalier, M.; Zarzo-Arias, A.; Guélat, J.; Mateo, R.G.; Guisan, A. Accounting for niche truncation to improve spatial and temporal predictions of species distributions. Front. Ecol. Evol. 2022, 10, 944116. [Google Scholar] [CrossRef]
- Li, Y.-P.; Gao, X.; An, Q.; Sun, Z.; Wang, H.-B. Ecological niche modeling based on ensemble algorithms to predicting current and future potential distribution of African swine fever virus in China. Sci. Rep. 2022, 12, 15614. [Google Scholar] [CrossRef]
- Amdouni, J.; Conte, A.; Ippoliti, C.; Candeloro, L.; Tora, S.; Sghaier, S.; Ben Hassine, T.; Fakhfekh, E.A.; Savini, G.; Hammami, S. Culex pipiens distribution in Tunisia: Identification of suitable areas through Random Forest and MaxEnt approaches. Vet. Med. Sci. 2022, 8, 2703–2715. [Google Scholar] [CrossRef]
- Andreo, V.; Cuervo, P.F.; Porcasi, X.; Lopez, L.; Guzman, C.; Scavuzzo, C.M. Towards a workflow for operational mapping of Aedes aegypti at urban scale based on remote sensing. Remote Sens. Appl. Soc. Environ. 2021, 23, 100554. [Google Scholar] [CrossRef]
- Hahn, M.B.; Feirer, S.; Monaghan, A.J.; Lane, R.S.; Eisen, R.J.; Padgett, K.A.; Kelly, M. Modeling future climate suitability for the western blacklegged tick, Ixodes pacificus, in California with an emphasis on land access and ownership. Ticks Tick-Borne Dis. 2021, 12, 101789. [Google Scholar] [CrossRef]
- Lippi, C.A.; Gaff, H.D.; White, A.L.; John, H.K.S.; Richards, A.L.; Ryan, S.J. Exploring the Niche of Rickettsia montanensis (Rickettsiales: Rickettsiaceae) Infection of the American Dog Tick (Acari: Ixodidae), Using Multiple Species Distribution Model Approaches. J. Med. Entomol. 2021, 58, 1083–1092. [Google Scholar] [CrossRef]
- Moo-Llanes, D.; López-Ordóñez, T.; Torres-Monzón, J.; Mosso-González, C.; Casas-Martínez, M.; Samy, A. Assessing the Potential Distributions of the Invasive Mosquito Vector Aedes albopictus and Its Natural Wolbachia Infections in México. Insects 2021, 12, 143. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, M.; Broennimann, O.; Cornuault, J.; Guisan, A. Data integration methods to account for spatial niche truncation effects in regional projections of species distribution. Ecol. Appl. 2021, 31, e02427. [Google Scholar] [CrossRef]
- Tjaden, N.; Cheng, Y.; Beierkuhnlein, C.; Thomas, S. Chikungunya Beyond the Tropics: Where and When Do We Expect Disease Transmission in Europe? Viruses 2021, 13, 1024. [Google Scholar] [CrossRef]
- Tjaden, N.B.; Suk, J.E.; Fischer, D.; Thomas, S.M.; Beierkuhnlein, C.; Semenza, J.C. Modelling the effects of global climate change on Chikungunya transmission in the 21st century. Sci. Rep. 2017, 7, 3813. [Google Scholar] [CrossRef]
- Ochida, N.; Mangeas, M.; Dupont-Rouzeyrol, M.; Dutheil, C.; Forfait, C.; Peltier, A.; Descloux, E.; Menkes, C. Modeling present and future climate risk of dengue outbreak, a case study in New Caledonia. Environ. Health 2022, 21, 20. [Google Scholar] [CrossRef] [PubMed]
- Cunze, S.; Kochmann, J.; Klimpel, S. Global occurrence data improve potential distribution models for Aedes japonicus japonicus in non-native regions. Pest Manag. Sci. 2020, 76, 1814–1822. [Google Scholar] [CrossRef] [PubMed]
- Yañez-Arenas, C.; Rioja-Nieto, R.; A Martín, G.; Dzul-Manzanilla, F.; Chiappa-Carrara, X.; Buenfil-Ávila, A.; Manrique-Saide, P.; Morales, F.C.; Díaz-Quiñónez, J.A.; Pérez-Rentería, C.; et al. Characterizing environmental suitability of Aedes albopictus (Diptera: Culicidae) in Mexico based on regional and global niche models. J. Med. Entomol. 2018, 55, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Echeverry-Cárdenas, E.; López-Castañeda, C.; Carvajal-Castro, J.D.; Aguirre-Obando, O.A. Potential geographic distribution of the tiger mosquito Aedes albopictus (Skuse, 1894) (Diptera: Culicidae) in current and future conditions for Colombia. PLoS Negl. Trop. Dis. 2021, 15, e0008212. [Google Scholar] [CrossRef] [PubMed]
- Titeux, N.; Maes, D.; Van Daele, T.; Onkelinx, T.; Heikkinen, R.K.; Romo, H.; García-Barros, E.; Munguira, M.L.; Thuiller, W.; Van Swaay, C.A.M.; et al. The need for large-scale distribution data to estimate regional changes in species richness under future climate change. Divers. Distrib. 2017, 23, 1393–1407. [Google Scholar] [CrossRef]
- Carretero, M.A.; Sillero, N. Evaluating how species niche modelling is affected by partial distributions with an empirical case. Acta Oecol. 2016, 77, 207–216. [Google Scholar] [CrossRef]
- Phillips, S.J.; Elith, J. On estimating probability of presence from use–availability or presence–background data. Ecology 2013, 94, 1409–1419. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A., Jr. A practical guide to MaxEnt for modeling species’ distributions: What it does, and why inputs and settings matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- Anderson, R.P.; Raza, A. The effect of the extent of the study region on GIS models of species geographic distributions and estimates of niche evolution: Preliminary tests with montane rodents (genus Nephelomys) in Venezuela. J. Biogeogr. 2010, 37, 1378–1393. [Google Scholar] [CrossRef]
- Nekola, J.C.; Divíšek, J.; Horsák, M. The nature of dispersal barriers and their impact on regional species pool richness and turnover. Glob. Ecol. Biogeogr. 2022, 31, 1470–1500. [Google Scholar] [CrossRef]
- Pang, S.E.H.; Zeng, Y.; Alban, J.D.T.; Webb, E.L. Occurrence–habitat mismatching and niche truncation when modelling distributions affected by anthropogenic range contractions. Divers. Distrib. 2022, 28, 1327–1343. [Google Scholar] [CrossRef]
- Machado-Stredel, F.; Cobos, M.E.; Peterson, A.T. A simulation-based method for selecting calibration areas for ecological niche models and species distribution models. Front. Biogeogr. 2021, 13, e48814. [Google Scholar] [CrossRef]
- Escalante, T. Un ensayo sobre regionalización biogeográfica. Rev. Mex. Biodivers. 2009, 80, 551–560. [Google Scholar] [CrossRef]
- Morrone, J.J. The spectre of biogeographical regionalization. J. Biogeogr. 2018, 45, 282–288. [Google Scholar] [CrossRef]
- Sierra-Morales, P.; Rojas-Soto, O.; Ríos-Muñoz, C.A.; Ochoa-Ochoa, L.M.; Flores-Rodríguez, P.; Almazán-Núñez, R.C. Climate change projections suggest severe decreases in the geographic ranges of bird species restricted to Mexican humid mountain forests. Glob. Ecol. Conserv. 2021, 30, e01794. [Google Scholar] [CrossRef]
- Ramsey, J.M.; Peterson, A.T.; Carmona-Castro, O.; Moo-Llanes, D.A.; Nakazawa, Y.; Butrick, M.; Tun-Ku, E.; De La Cruz-Félix, K.; Ibarra-Cerdeña, C.N. Atlas of Mexican Triatominae (Reduviidae: Hemiptera) and vector transmission of Chagas disease. Mem. Inst. Oswaldo Cruz 2015, 110, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Soberón, J.M. Niche and area of distribution modeling: A population ecology perspective. Ecography 2010, 33, 159–167. [Google Scholar] [CrossRef]
- Bystriakova, N.; Peregrym, M.; Erkens, R.H.; Bezsmertna, O.; Schneider, H. Sampling bias in geographic and environmental space and its effect on the predictive power of species distribution models. Syst. Biodivers. 2012, 10, 305–315. [Google Scholar] [CrossRef]
- Hughes, A.C.; Orr, M.C.; Ma, K.; Costello, M.J.; Waller, J.; Provoost, P.; Yang, Q.; Zhu, C.; Qiao, H. Sampling biases shape our view of the natural world. Ecography 2021, 44, 1259–1269. [Google Scholar] [CrossRef]
- Rocha-Ortega, M.; Rodriguez, P.; Córdoba-Aguilar, A. Geographical, temporal and taxonomic biases in insect GBIF data on biodiversity and extinction. Ecol. Entomol. 2021, 46, 718–728. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Elith, J.; Graham, C.H.; Lehmann, A.; Leathwick, J.; Ferrier, S. Sample selection bias and presence-only distribution models: Implications for background and pseudo-absence data. Ecol. Appl. 2009, 19, 181–197. [Google Scholar] [CrossRef]
- Laporta, G.Z.; Potter, A.M.; Oliveira, J.F.A.; Bourke, B.P.; Pecor, D.B.; Linton, Y.-M. Global Distribution of Aedes aegypti and Aedes albopictus in a Climate Change Scenario of Regional Rivalry. Insects 2023, 14, 49. [Google Scholar] [CrossRef]
- Aiello-Lammens, M.E.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Anderson, R.P. spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 2015, 38, 541–545. [Google Scholar] [CrossRef]
- Castellanos, A.A.; Huntley, J.W.; Voelker, G.; Lawing, A.M. Environmental filtering improves ecological niche models across multiple scales. Methods Ecol. Evol. 2019, 10, 481–492. [Google Scholar] [CrossRef]
- Kramer-Schadt, S.; Niedballa, J.; Pilgrim, J.D.; Schröder, B.; Lindenborn, J.; Reinfelder, V.; Stillfried, M.; Heckmann, I.; Scharf, A.K.; Augeri, D.M.; et al. The importance of correcting for sampling bias in MaxEnt species distribution models. Divers. Distrib. 2013, 19, 1366–1379. [Google Scholar] [CrossRef]
- Boria, R.A.; Olson, L.E.; Goodman, S.M.; Anderson, R.P. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol. Model. 2014, 275, 73–77. [Google Scholar] [CrossRef]
- Varela, S.; Anderson, R.P.; García-Valdés, R.; Fernández González, F. Environmental filters reduce the effects of sampling bias and improve predictions of ecological niche models. Ecography 2014, 37, 1084–1091. [Google Scholar] [CrossRef]
- Cosentino, F.; Maiorano, L. Is geographic sampling bias representative of environmental space? Ecol. Inform. 2021, 64, 101369. [Google Scholar] [CrossRef]
- Barber, R.A.; Ball, S.G.; Morris, R.K.A.; Gilbert, F. Target-group backgrounds prove effective at correcting sampling bias in Maxent models. Divers. Distrib. 2022, 28, 128–141. [Google Scholar] [CrossRef]
- Vollering, J.; Halvorsen, R.; Auestad, I.; Rydgren, K. Bunching up the background betters bias in species distribution models. Ecography 2019, 42, 1717–1727. [Google Scholar] [CrossRef]
- De Oliveira, G.; Rangel, T.F.; Lima-Ribeiro, M.S.; Terribile, L.C.; Diniz-Filho, J.A.F. Evaluating, partitioning, and mapping the spatial autocorrelation component in ecological niche modeling: A new approach based on environmentally equidistant records. Ecography 2014, 37, 637–647. [Google Scholar] [CrossRef]
- Peterson, A.T.; Cobos, M.E.; Jiménez-García, D. Major challenges for correlational ecological niche model projections to future climate conditions. Ann. N. Y. Acad. Sci. 2018, 1429, 66–77. [Google Scholar] [CrossRef]
- Warren, D.L.; Seifert, S.N. Ecological niche modeling in Maxent: The importance of model complexity and the performance of model selection criteria. Ecol. Appl. 2011, 21, 335–342. [Google Scholar] [CrossRef]
- Peterson, A.T.; Nakazawa, Y. Environmental data sets matter in ecological niche modelling: An example with Solenopsis invicta and Solenopsis richteri. Glob. Ecol. Biogeogr. 2008, 17, 135–144. [Google Scholar] [CrossRef]
- Cobos, M.E.; Peterson, A.T.; Osorio-Olvera, L.; Jiménez-García, D. An exhaustive analysis of heuristic methods for variable selection in ecological niche modeling and species distribution modeling. Ecol. Inform. 2019, 53, 100983. [Google Scholar] [CrossRef]
- Zurell, D.; Franklin, J.; König, C.; Bouchet, P.J.; Dormann, C.F.; Elith, J.; Fandos, G.; Feng, X.; Guillera-Arroita, G.; Guisan, A.; et al. A standard protocol for reporting species distribution models. Ecography 2020, 43, 1261–1277. [Google Scholar] [CrossRef]
- Muscarella, R.; Galante, P.J.; Soley-Guardia, M.; Boria, R.A.; Kass, J.M.; Uriarte, M.; Anderson, R.P. ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxentecological niche models. Methods Ecol. Evol. 2014, 5, 1198–1205. [Google Scholar] [CrossRef]
- Vignali, S.; Barras, A.G.; Arlettaz, R.; Braunisch, V. SDMtune: An R package to tune and evaluate species distribution models. Ecol. Evol. 2020, 10, 11488–11506. [Google Scholar] [CrossRef] [PubMed]
- Radosavljevic, A.; Anderson, R.P. Making better Maxent models of species distributions: Complexity, overfitting and evaluation. J. Biogeogr. 2014, 41, 629–643. [Google Scholar] [CrossRef]
- Kass, J.M.; Muscarella, R.; Galante, P.J.; Bohl, C.L.; Pinilla-Buitrago, G.E.; Boria, R.A.; Soley-Guardia, M.; Anderson, R.P. ENMeval 2.0: Redesigned for customizable and reproducible modeling of species’ niches and distributions. Methods Ecol. Evol. 2021, 12, 1602–1608. [Google Scholar] [CrossRef]
- Kass, J.M.; Vilela, B.; Aiello-Lammens, M.E.; Muscarella, R.; Merow, C.; Anderson, R.P. Wallace: A flexible platform for reproducible modeling of species niches and distributions built for community expansion. Methods Ecol. Evol. 2018, 9, 1151–1156. [Google Scholar] [CrossRef]
- Kass, J.M.; Pinilla-Buitrago, G.E.; Paz, A.; Johnson, B.A.; Grisales-Betancur, V.; Meenan, S.I.; Attali, D.; Broennimann, O.; Galante, P.J.; Maitner, B.S.; et al. wallace 2: A shiny app for modeling species niches and distributions redesigned to facilitate expansion via module contributions. Ecography 2023, 2023, e06547. [Google Scholar] [CrossRef]
- Cobos, M.E.; Townsend Peterson, A.; Barve, N.; Osorio-Olvera, L. kuenm: An R package for detailed development of ecological niche models using Maxent. PeerJ 2019, 7, e6281. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.A.; Stralberg, D.; Jongsomjit, D.; Howell, C.A.; Snyder, M.A. Niches, models, and climate change: Assessing the assumptions and uncertainties. Proc. Natl. Acad. Sci. USA 2009, 106 (Suppl. S2), 19729–19736. [Google Scholar] [CrossRef]
- Pearson, R.G.; Thuiller, W.; Araújo, M.B.; Martínez-Meyer, E.; Brotons, L.; McClean, C.; Miles, L.; Segurado, P.; Dawson, T.; Lees, D.C. Model-based uncertainty in species range prediction. J. Biogeogr. 2006, 33, 1704–1711. [Google Scholar] [CrossRef]
- Alkishe, A.; Cobos, M.E.; Peterson, A.T.; Samy, A.M. Recognizing sources of uncertainty in disease vector ecological niche models: An example with the tick Rhipicephalus sanguineus sensu lato. Perspect. Ecol. Conserv. 2020, 18, 91–102. [Google Scholar] [CrossRef]
- Qiao, H.; Feng, X.; Escobar, L.E.; Peterson, A.T.; Soberón, J.; Zhu, G.; Papeş, M. An evaluation of transferability of ecological niche models. Ecography 2019, 42, 521–534. [Google Scholar] [CrossRef]
- Simoes, M.; Romero-Alvarez, D.; Nuñez-Penichet, C.; Jiménez, L.; Cobos, M.E. General Theory and Good Practices in Ecological Niche Modeling: A Basic Guide. Biodivers. Inform. 2020, 15, 67–68. [Google Scholar] [CrossRef]
- Elith, J.; Kearney, M.; Phillips, S. The art of modelling range-shifting species. Methods Ecol. Evol. 2010, 1, 330–342. [Google Scholar] [CrossRef]
- Witmer, F.D.W.; Nawrocki, T.W.; Hahn, M. Modeling Geographic Uncertainty in Current and Future Habitat for Potential Populations of Ixodes pacificus (Acari: Ixodidae) in Alaska. J. Med. Entomol. 2022, 59, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Yates, K.L.; Bouchet, P.J.; Caley, M.J.; Mengersen, K.; Randin, C.F.; Parnell, S.; Fielding, A.H.; Bamford, A.J.; Ban, S.; Barbosa, A.M.; et al. Outstanding Challenges in the Transferability of Ecological Models. Trends Ecol. Evol. 2018, 33, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Alkhamis, M.A.; Fountain-Jones, N.M.; Aguilar-Vega, C.; Sánchez-Vizcaíno, J.M. Environment, vector, or host? Using machine learning to untangle the mechanisms driving arbovirus outbreaks. Ecol. Appl. 2021, 31, e02407. [Google Scholar] [CrossRef]
- Peterson, A.T. Ecologic Niche Modeling and Spatial Patterns of Disease Transmission. Emerg. Infect. Dis. 2006, 12, 1822–1826. [Google Scholar] [CrossRef]
- Lockwood, J.L.; Hoopes, M.F.; Marchetti, M.P. Invasion Ecology, 2nd ed.; Wiley-Blackwell Publishing: Oxford, UK, 2013; p. 444. [Google Scholar]
- Wilson, J.R.; Dormontt, E.E.; Prentis, P.J.; Lowe, A.J.; Richardson, D.M. Something in the way you move: Dispersal pathways affect invasion success. Trends Ecol. Evol. 2009, 24, 136–144. [Google Scholar] [CrossRef]
- Crowl, T.A.; Crist, T.O.; Parmenter, R.R.; Belovsky, G.; Lugo, A.E. The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 2008, 6, 238–246. [Google Scholar] [CrossRef]
- Jimenezvalverde, A.; Peterson, A.T.; Soberon, J.; Overton, J.M.; Aragón, P.; Lobo, J.M. Use of niche models in invasive species risk assessments. Biol. Invasions 2011, 13, 2785–2797. [Google Scholar] [CrossRef]
- Castaño-Quintero, S.; Escobar-Luján, J.; Osorio-Olvera, L.; Peterson, A.T.; Chiappa-Carrara, X.; Martínez-Meyer, E.; Yañez-Arenas, C. Supraspecific units in correlative niche modeling improves the prediction of geographic potential of biological invasions. PeerJ 2020, 8, e10454. [Google Scholar] [CrossRef]
- Sanderson, B.M.; Knutti, R.; Caldwell, P. A Representative Democracy to Reduce Interdependency in a Multimodel Ensemble. J. Clim. 2015, 28, 5171–5194. [Google Scholar] [CrossRef]
- Chen, I.-C.; Hill, J.K.; Ohlemüller, R.; Roy, D.B.; Thomas, C.D. Rapid Range Shifts of Species Associated with High Levels of Climate Warming. Science 2011, 333, 1024–1026. [Google Scholar] [CrossRef] [PubMed]
- Pili, A.N.; Tingley, R.; Sy, E.Y.; Diesmos, M.L.L.; Diesmos, A.C. Niche shifts and environmental non-equilibrium undermine the usefulness of ecological niche models for invasion risk assessments. Sci. Rep. 2020, 10, 7972. [Google Scholar] [CrossRef]
- Chuang, A.; Peterson, C.R. Expanding population edges: Theories, traits, and trade-offs. Glob. Chang. Biol. 2016, 22, 494–512. [Google Scholar] [CrossRef] [PubMed]
- Arenas, M.; Ray, N.; Currat, M.; Excoffier, L. Consequences of Range Contractions and Range Shifts on Molecular Diversity. Mol. Biol. Evol. 2012, 29, 207–218. [Google Scholar] [CrossRef]
- Cunze, S.; Glock, G.; Kochmann, J.; Klimpel, S. Ticks on the move—Climate change-induced range shifts of three tick species in Europe: Current and future habitat suitability for Ixodes ricinus in comparison with Dermacentor reticulatus and Dermacentor marginatus. Parasitol. Res. 2022, 121, 2241–2252. [Google Scholar] [CrossRef]
- Flenniken, J.M.; Tuten, H.C.; Vineer, H.R.; Phillips, V.C.; Stone, C.M.; Allan, B.F. Environmental Drivers of Gulf Coast Tick (Acari: Ixodidae) Range Expansion in the United States. J. Med. Entomol. 2022, 59, 1625–1635. [Google Scholar] [CrossRef]
- Alkishe, A.; Raghavan, R.K.; Peterson, A.T. Likely Geographic Distributional Shifts among Medically Important Tick Species and Tick-Associated Diseases under Climate Change in North America: A Review. Insects 2021, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Steger, J.; Schneider, A.; Brandl, R.; Hotes, S. Effects of projected climate change on the distribution of Mantis religiosa suggest expansion followed by contraction. Web Ecol. 2020, 20, 107–115. [Google Scholar] [CrossRef]
- McIntyre, S.; Rangel, E.F.; Ready, P.D.; Carvalho, B.M. Species-specific ecological niche modelling predicts different range contractions for Lutzomyia intermedia and a related vector of Leishmania braziliensis following climate change in South America. Parasites Vectors 2017, 10, 157. [Google Scholar] [CrossRef]
- Wiens, J.J. Species Delimitation: New Approaches for Discovering Diversity. Syst. Biol. 2007, 56, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Serrano, D.F.; Knowles, L.L. Ecological niche models in phylogeographic studies: Applications, advances and precautions. Mol. Ecol. Resour. 2014, 14, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J.; Graham, C.H. Niche Conservatism: Integrating Evolution, Ecology, and Conservation Biology. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 519–539. [Google Scholar] [CrossRef]
- Foley, D.H.; Rueda, L.M.; Peterson, A.T.; Wilkerson, R.C. Potential Distribution of Two Species in the Medically Important Anopheles minimus Complex (Diptera: Culicidae). J. Med. Entomol. 2008, 45, 852–860. [Google Scholar] [CrossRef]
- Collart, F.; Hedenäs, L.; Broennimann, O.; Guisan, A.; Vanderpoorten, A. Intraspecific differentiation: Implications for niche and distribution modelling. J. Biogeogr. 2021, 48, 415–426. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuervo, P.F.; Artigas, P.; Lorenzo-Morales, J.; Bargues, M.D.; Mas-Coma, S. Ecological Niche Modelling Approaches: Challenges and Applications in Vector-Borne Diseases. Trop. Med. Infect. Dis. 2023, 8, 187. https://doi.org/10.3390/tropicalmed8040187
Cuervo PF, Artigas P, Lorenzo-Morales J, Bargues MD, Mas-Coma S. Ecological Niche Modelling Approaches: Challenges and Applications in Vector-Borne Diseases. Tropical Medicine and Infectious Disease. 2023; 8(4):187. https://doi.org/10.3390/tropicalmed8040187
Chicago/Turabian StyleCuervo, Pablo Fernando, Patricio Artigas, Jacob Lorenzo-Morales, María Dolores Bargues, and Santiago Mas-Coma. 2023. "Ecological Niche Modelling Approaches: Challenges and Applications in Vector-Borne Diseases" Tropical Medicine and Infectious Disease 8, no. 4: 187. https://doi.org/10.3390/tropicalmed8040187
APA StyleCuervo, P. F., Artigas, P., Lorenzo-Morales, J., Bargues, M. D., & Mas-Coma, S. (2023). Ecological Niche Modelling Approaches: Challenges and Applications in Vector-Borne Diseases. Tropical Medicine and Infectious Disease, 8(4), 187. https://doi.org/10.3390/tropicalmed8040187









