Production and Immunological Characterization of scFv Specific to Epitope of Opisthorchis viverrini Rhophilin-Associated Tail Protein 1-like (OvROPN1L)
Abstract
1. Introduction
2. Materials and Methods
2.1. Production of Recombinant Protein OvROPN1L
2.2. Polyclonal Antibody Production
2.3. Preparation of Coproantigens
2.4. Immobilization of Antigens
2.5. Biopanning
2.6. Screening of the scFv Phages Binding with Full-Length rOvROPN1L and Non-Infected Coproantigen
2.7. Production of Soluble Anti-OvROPN1L scFv Antibody
2.8. Binding Assessment of Anti-OvROPN1L scFv Antibody with Coproantigens
2.9. Molecular Modeling and Docking
2.10. Statistical Analysis
3. Results
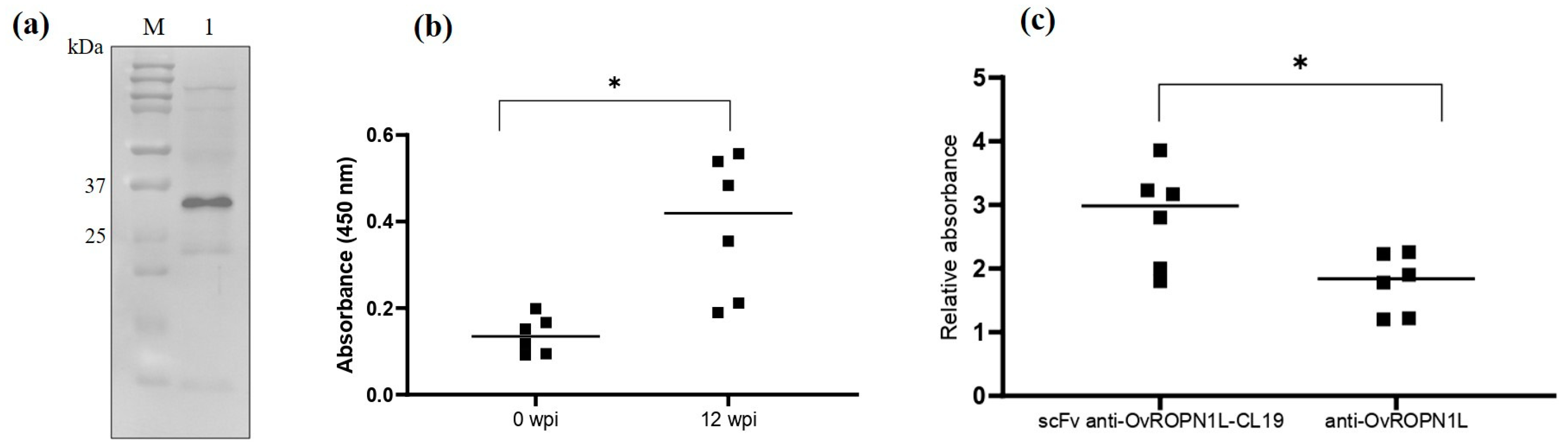
3.1. scFv Phages Specifically Bound to Recombinant OvROPN1L
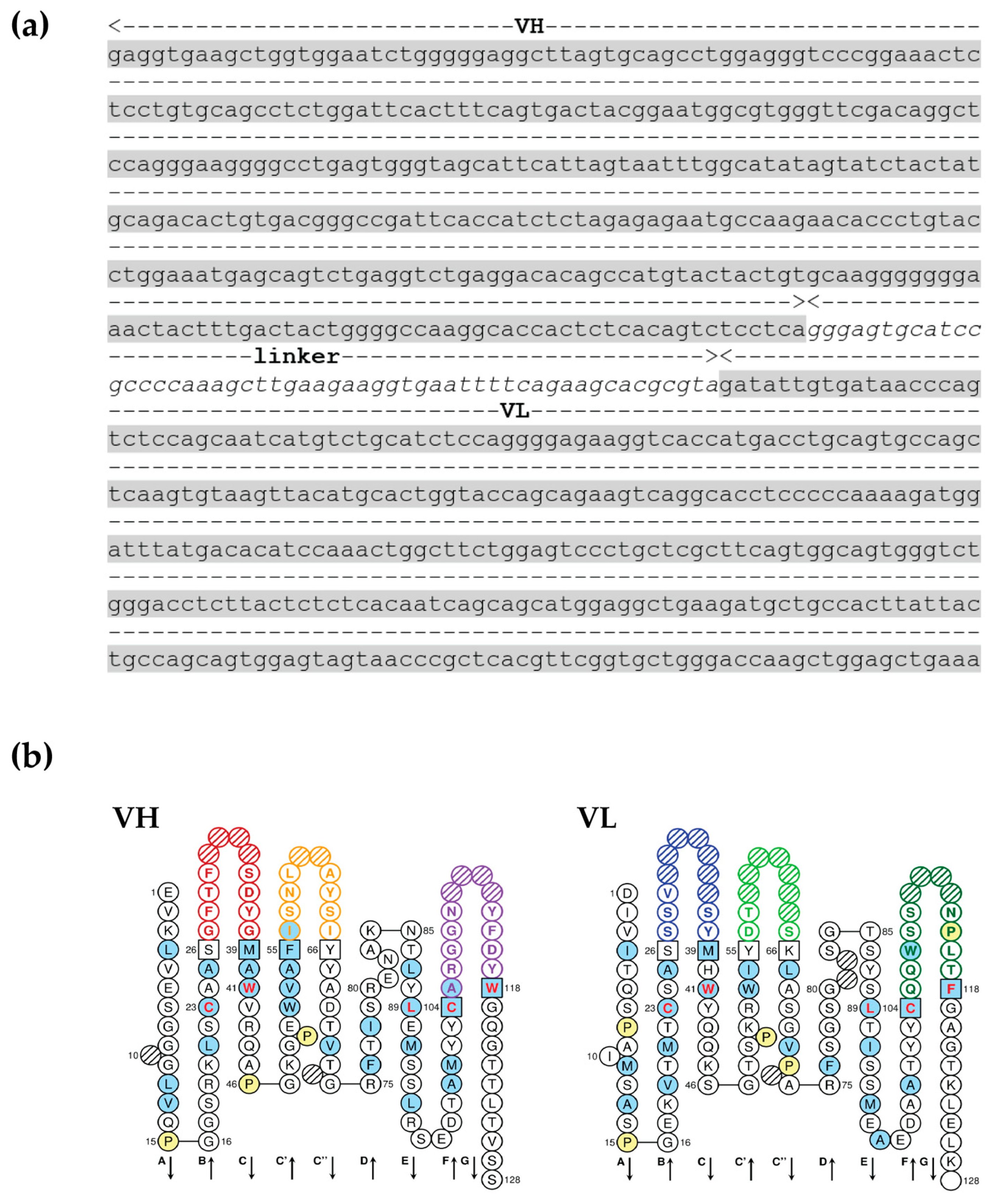
3.2. scFv Specific to OvROPN1L Has Been Successfully Cloned
3.3. Soluble Recombinant scFv Anti-OvROPN1L-CL19 Was Successfully Produced and Preliminarily Tested against Coproantigens
3.4. Molecular Docking Confirmed the Binding of scFv Anti-OvROPN1L-CL19 and OvROPN1L
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sripa, B.; Kaewkes, S.; Sithithaworn, P.; Mairiang, E.; Laha, T.; Smout, M.; Pairojkul, C.; Bhudhisawasdi, V.; Tesana, S.; Thinkamrop, B.; et al. Liver Fluke Induces Cholangiocarcinoma. PLoS Med. 2007, 4, e201. [Google Scholar] [CrossRef] [PubMed]
- Sripa, B.; Suwannatrai, A.T.; Sayasone, S.; Do, D.T.; Khieu, V.; Yang, Y. Current status of human liver fluke infections in the Greater Mekong Subregion. Acta Trop. 2021, 224, 106133. [Google Scholar] [CrossRef] [PubMed]
- Sanpool, O.; Aung, W.P.P.; Rodpai, R.; Maleewong, W.; Intapan, P.M. Human liver fluke Opisthorchis viverrini (Trematoda, Opisthorchiidae) in Central Myanmar: New records of adults and metacercariae identified by morphology and molecular analysis. Acta Trop. 2018, 185, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Crellen, T.; Sithithaworn, P.; Pitaksakulrat, O.; Khuntikeo, N.; Medley, G.F.; Hollingsworth, T.D. Towards Evidence-based Control of Opisthorchis viverrini. Trends Parasitol. 2021, 37, 370–380. [Google Scholar] [CrossRef]
- Johansen, M.V.; Sithithaworn, P.; Bergquist, R.; Utzinger, J. Towards improved diagnosis of zoonotic trematode infections in Southeast Asia. Adv. Parasitol. 2010, 73, 171–195. [Google Scholar] [CrossRef] [PubMed]
- Johansen, M.V.; Lier, T.; Sithithaworn, P. Towards improved diagnosis of neglected zoonotic trematodes using a One Health approach. Acta Trop. 2015, 141, 161–169. [Google Scholar] [CrossRef]
- Charoensuk, L.; Subrungruang, I.; Mungthin, M.; Pinlaor, S.; Suwannahitatorn, P. Comparison of stool examination techniques to detect Opisthorchis viverrini in low intensity infection. Acta Trop. 2019, 191, 13–16. [Google Scholar] [CrossRef]
- Kopolrat, K.Y.; Singthong, S.; Khuntikeo, N.; Loilome, W.; Worasith, C.; Homwong, C.; Wangboon, C.; Yasaka, P.; Eamudomkarn, C.; Pitaksakulrat, O.; et al. Performance of Mini Parasep(®) SF stool concentrator kit, Kato-Katz, and formalin-ethyl acetate concentration methods for diagnosis of opisthorchiasis in Northeast Thailand. Parasites Vectors 2022, 15, 234. [Google Scholar] [CrossRef]
- Phadungsil, W.; Pumpa, S.; Sirisabhabhorn, K.; Geadkaew-Krenc, A.; Grams, R.; Mungthin, M.; Ruang-Areerate, T.; Adisakwattana, P.; Labbunruang, N.; Martviset, P. Efficiency of the Stool-PCR Test Targeting NADH Dehydrogenase (Nad) Subunits for Detection of Opisthorchis viverrini Eggs. J. Trop. Med. 2021, 2021, 3957545. [Google Scholar] [CrossRef]
- Xue, Y.P.; Kao, M.C.; Lan, C.Y. Novel mitochondrial complex I-inhibiting peptides restrain NADH dehydrogenase activity. Sci. Rep. 2019, 9, 13694. [Google Scholar] [CrossRef]
- Kerscher, S.J. Diversity and origin of alternative NADH:ubiquinone oxidoreductases. Biochim. Biophys. Acta (BBA)-Bioenerg. 2000, 1459, 274–283. [Google Scholar] [CrossRef]
- Buathong, S.; Leelayoova, S.; Mungthin, M.; Ruang-areerate, T.; Naaglor, T.; Suwannahitatorn, P.; Piyaraj, P.; Taamasri, P.; Tan-ariya, P. Molecular discrimination of Opisthorchis-like eggs from residents in a rural community of central Thailand. PLoS Negl. Trop. Dis. 2017, 11, e0006030. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Biswal, D.K.; Roy, B.; Tandon, V. Molecular characterization of Opisthorchis noverca (Digenea: Opisthorchiidae) based on nuclear ribosomal ITS2 and mitochondrial COI genes. J. Helminthol. 2016, 90, 607–614. [Google Scholar] [CrossRef]
- Pumpa, S.; Phadungsil, W.; Grams, R.; Martviset, P.; Ruang-Areerate, T.; Mungthin, M.; Geadkaew-Krenc, A. Improvement of a PCR-based method for the detection of Opisthorchis viverrini eggs in human stool samples by targeting internal transcribed spacer-2 (ITS-2), cytochrome oxidase subunit 1 (cox1), and cytochrome b (cyb). J. Parasit. Dis. 2021, 45, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Buathong, S.; Leelayoova, S.; Mungthin, M.; Naaglor, T.; Taamasri, P.; Suwannahitatorn, P.; Tan-Ariya, P. Development and evaluation of PCR methods based on cytochrome c oxidase subunit one (cox1) and NADH dehydrogenase subunit one gene (nad1) to detect Opisthorchis viverrini in human fecal samples. Parasitol. Res. 2015, 114, 3547–3549. [Google Scholar] [CrossRef]
- Cai, X.Q.; Yu, H.Q.; Li, R.; Yue, Q.Y.; Liu, G.H.; Bai, J.S.; Deng, Y.; Qiu, D.Y.; Zhu, X.Q. Rapid detection and differentiation of Clonorchis sinensis and Opisthorchis viverrini using real-time PCR and high resolution melting analysis. Sci. World J. 2014, 2014, 893981. [Google Scholar] [CrossRef]
- Arimatsu, Y.; Kaewkes, S.; Laha, T.; Hong, S.J.; Sripa, B. Rapid detection of Opisthorchis viverrini copro-DNA using loop-mediated isothermal amplification (LAMP). Parasitol. Int. 2012, 61, 178–182. [Google Scholar] [CrossRef]
- Wongratanacheewin, S.; Sermswan, R.W.; Sirisinha, S. Immunology and molecular biology of Opisthorchis viverrini infection. Acta Trop. 2003, 88, 195–207. [Google Scholar] [CrossRef]
- Sirisinha, S.; Chawengkirttikul, R.; Sermswan, R. Immunodiagnosis of opisthorchiasis. Southeast Asian J. Trop. Med. Public Health 1991, 22, 179–183. [Google Scholar]
- Laha, T.; Sripa, J.; Sripa, B.; Pearson, M.; Tribolet, L.; Kaewkes, S.; Sithithaworn, P.; Brindley, P.J.; Loukas, A. Asparaginyl endopeptidase from the carcinogenic liver fluke, Opisthorchis viverrini, and its potential for serodiagnosis. Int. J. Infect. Dis. 2008, 12, e49–e59. [Google Scholar] [CrossRef]
- Teimoori, S.; Arimatsu, Y.; Laha, T.; Kaewkes, S.; Sereerak, P.; Tangkawattana, S.; Brindley, P.J.; Sripa, B. Immunodiagnosis of opisthorchiasis using parasite cathepsin F. Parasitol. Res. 2015, 114, 4571–4578. [Google Scholar] [CrossRef] [PubMed]
- Akai, P.S.; Pungpak, S.; Chaicumpa, W.; Viroj, K.; Bunnag, D.; Befus, A.D. Serum antibody response to Opisthorchis viverrini antigen as a marker for opisthorchiasis-associated cholangiocarcinoma. Trans. R. Soc. Trop. Med. Hyg. 1994, 88, 471–474. [Google Scholar] [CrossRef] [PubMed]
- Worasith, C.; Kamamia, C.; Yakovleva, A.; Duenngai, K.; Wangboon, C.; Sithithaworn, J.; Watwiengkam, N.; Namwat, N.; Techasen, A.; Loilome, W.; et al. Advances in the Diagnosis of Human Opisthorchiasis: Development of Opisthorchis viverrini Antigen Detection in Urine. PLoS Negl. Trop. Dis. 2015, 9, e0004157. [Google Scholar] [CrossRef] [PubMed]
- Sadaow, L.; Rodpai, R.; Janwan, P.; Boonroumkaew, P.; Sanpool, O.; Thanchomnang, T.; Yamasaki, H.; Ittiprasert, W.; Mann, V.H.; Brindley, P.J.; et al. An Innovative Test for the Rapid Detection of Specific IgG Antibodies in Human Whole-Blood for the Diagnosis of Opisthorchis viverrini Infection. Trop. Med. Infect. Dis. 2022, 7, 308. [Google Scholar] [CrossRef] [PubMed]
- Sripa, J.; Brindley, P.J.; Sripa, B.; Loukas, A.; Kaewkes, S.; Laha, T. Evaluation of liver fluke recombinant cathepsin B-1 protease as a serodiagnostic antigen for human opisthorchiasis. Parasitol. Int. 2012, 61, 191–195. [Google Scholar] [CrossRef]
- Ruangsittichai, J.; Viyanant, V.; Vichasri-Grams, S.; Sobhon, P.; Tesana, S.; Upatham, E.S.; Hofmann, A.; Korge, G.; Grams, R. Opisthorchis viverrini: Identification of a glycine–tyrosine rich eggshell protein and its potential as a diagnostic tool for human opisthorchiasis. Int. J. Parasitol. 2006, 36, 1329–1339. [Google Scholar] [CrossRef]
- Goździk, K.; Engström, A.; Höglund, J. Optimization of in-house ELISA based on recombinant major sperm protein (rMSP) of Dictyocaulus viviparus for the detection of lungworm infection in cattle. Res. Vet. Sci. 2012, 93, 813–818. [Google Scholar] [CrossRef]
- Carr, D.W.; Fujita, A.; Stentz, C.L.; Liberty, G.A.; Olson, G.E.; Narumiya, S. Identification of Sperm-specific Proteins That Interact with A-kinase Anchoring Proteins in a Manner Similar to the Type II Regulatory Subunit of PKA*. J. Biol. Chem. 2001, 276, 17332–17338. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Wei, B.; Lai, Y.; Yan, Q.; Gui, Y.; Cai, Z. Functional expression of ropporin in human testis and ejaculated spermatozoa. J. Androl. 2011, 32, 26–32. [Google Scholar] [CrossRef]
- Pelloni, M.; Paoli, D.; Majoli, M.; Pallotti, F.; Carlini, T.; Lenzi, A.; Lombardo, F. Molecular study of human sperm RNA: Ropporin and CABYR in asthenozoospermia. J. Endocrinol. Investig. 2018, 41, 781–787. [Google Scholar] [CrossRef]
- Peck, J.W.; Oberst, M.; Bouker, K.B.; Bowden, E.; Burbelo, P.D. The RhoA-binding protein, rhophilin-2, regulates actin cytoskeleton organization. J. Biol. Chem. 2002, 277, 43924–43932. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Huang, X.; Li, Q.; He, L.; Li, S.; Chen, X.; Ouyang, Y.; Wang, X.; Lin, C. Rhophilin-associated tail protein 1 promotes migration and metastasis in triple negative breast cancer via activation of RhoA. Faseb. J. 2020, 34, 9959–9971. [Google Scholar] [CrossRef] [PubMed]
- Fujita, A.; Nakamura, K.; Kato, T.; Watanabe, N.; Ishizaki, T.; Kimura, K.; Mizoguchi, A.; Narumiya, S. Ropporin, a sperm-specific binding protein of rhophilin, that is localized in the fibrous sheath of sperm flagella. J. Cell Sci. 2000, 113, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Geadkaew-Krenc, A.; Grams, R.; Phadungsil, W.; Chaibangyang, W.; Kosa, N.; Adisakwattana, P.; Dekumyoy, P. Evaluation of Rhophilin Associated Tail Protein (ROPN1L) in the Human Liver Fluke Opisthorchis viverrini for Diagnostic Approach. Korean J. Parasitol. 2020, 58, 475–479. [Google Scholar] [CrossRef]
- Rattanachan, S.; Grams, R.; Tesana, S.; Smooker, P.M.; Grams, S.V. Opisthorchis viverrini: Analysis of the sperm-specific rhophilin associated tail protein 1-like. Acta Trop. 2014, 140, 34–40. [Google Scholar] [CrossRef]
- Geadkaew, A.; Kosa, N.; Siricoon, S.; Grams, S.V.; Grams, R. A 170kDa multi-domain cystatin of Fasciola gigantica is active in the male reproductive system. Mol. Biochem. Parasitol. 2014, 196, 100–107. [Google Scholar] [CrossRef]
- Martviset, P.; Chantree, P.; Chaimon, S.; Torungkitmangmi, N.; Prathaphan, P.; Ruangtong, J.; Sornchuer, P.; Thongsepee, N.; Sangpairoj, K.; Adisakwattana, P. Molecular Cloning and Characterization of a Fasciola gigantica Nuclear Receptor Subfamily 1 (FgNR1). Pathogens 2022, 11, 1458. [Google Scholar] [CrossRef]
- Mazidur Rahman, S.M.; Choi, M.H.; Bae, Y.M.; Hong, S.T. Coproantigen capture ELISA for detection of Clonorchis sinensis infection in experimentally infected rats. Parasitol. Int. 2012, 61, 203–207. [Google Scholar] [CrossRef]
- Thanongsaksrikul, J.; Srimanote, P.; Tongtawe, P.; Glab-Ampai, K.; Malik, A.A.; Supasorn, O.; Chiawwit, P.; Poovorawan, Y.; Chaicumpa, W. Identification and production of mouse scFv to specific epitope of enterovirus-71 virion protein-2 (VP2). Arch. Virol. 2018, 163, 1141–1152. [Google Scholar] [CrossRef]
- Giudicelli, V.; Duroux, P.; Kossida, S.; Lefranc, M.-P. IG and TR single chain fragment variable (scFv) sequence analysis: A new advanced functionality of IMGT/V-QUEST and IMGT/HighV-QUEST. BMC Immunol. 2017, 18, 35. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Vangone, A.; Bonvin, A.M. Contacts-based prediction of binding affinity in protein-protein complexes. eLife 2015, 4, e07454. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Phumrattanaprapin, W.; Pearson, M.; Pickering, D.; Tedla, B.; Smout, M.; Chaiyadet, S.; Brindley, P.J.; Loukas, A.; Laha, T. Monoclonal Antibodies Targeting an Opisthorchis viverrini Extracellular Vesicle Tetraspanin Protect Hamsters against Challenge Infection. Vaccines 2021, 9, 740. [Google Scholar] [CrossRef] [PubMed]
- Sirisinha, S.; Chawengkirttikul, R.; Sermswan, R.; Amornpant, S.; Mongkolsuk, S.; Panyim, S. Detection of Opisthorchis viverrini by Monoclonal Antibody-Based ELISA and DNA Hybridization. Am. J. Trop. Med. Hyg. 1991, 44, 140–145. [Google Scholar] [CrossRef]
- BILLINGS, P.B.; UTSAKHIT, N.; SIRISINHA, S. Monoclonal antibodies against Opisthorchis viverrini antigens. Parasite Immunol. 1990, 12, 545–557. [Google Scholar] [CrossRef]
- Chaicumpa, W.; Ybanez, L.; Kitikoon, V.; Pungpak, S.; Ruangkunaporn, Y.; Chongsa-nguan, M.; Sornmani, S. Detection of Opisthorchis viverrini antigens in stools using specific monoclonal antibody. Int. J. Parasitol. 1992, 22, 527–531. [Google Scholar] [CrossRef]
- Amornpunt, S.; Sarasombath, S.; Sirisinha, S. Production and characterization of monoclonal antibodies against the excretory-secretory antigen of the liver fluke (Opisthorchis viverrini). Int. J. Parasitol. 1991, 21, 421–428. [Google Scholar] [CrossRef]
- Sirisinha, S.; Chawengkirttikul, R.; Haswell-Elkins, M.R.; Elkins, D.B.; Kaewkes, S.; Sithithaworn, P. Evaluation of a Monoclonal Antibody-Based Enzyme-Linked Immunosorbent Assay for the Diagnosis of Opisthorchis viverrini Infection in an Endemic Area. Am. J. Trop. Med. Hyg. 1995, 52, 521–524. [Google Scholar] [CrossRef]
- Wongsaroj, T.; Sakolvaree, Y.; Chaicumpa, W.; Maleewong, W.; Kitikoon, V.; Tapchaisri, P.; Chongsa-nguan, M.; Cross, J.H. Affinity purified oval antigen for diagnosis of Opisthorchiasis viverrini. Asian Pac. J. Allergy Immunol. 2001, 19, 245–258. [Google Scholar] [PubMed]
- Worasith, C.; Wangboon, C.; Duenngai, K.; Kiatsopit, N.; Kopolrat, K.; Techasen, A.; Sithithaworn, J.; Khuntikeo, N.; Loilome, W.; Namwat, N.; et al. Comparing the performance of urine and copro-antigen detection in evaluating Opisthorchis viverrini infection in communities with different transmission levels in Northeast Thailand. PLoS Negl. Trop. Dis. 2019, 13, e0007186. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Thattanon, P.; Thanongsaksrikul, J.; Petvises, S.; Nathalang, O. Monoclonal antibody specific to the Di(a) blood group antigen generated by phage display technology. Blood Transfus. 2020, 18, 366–373. [Google Scholar] [CrossRef] [PubMed]

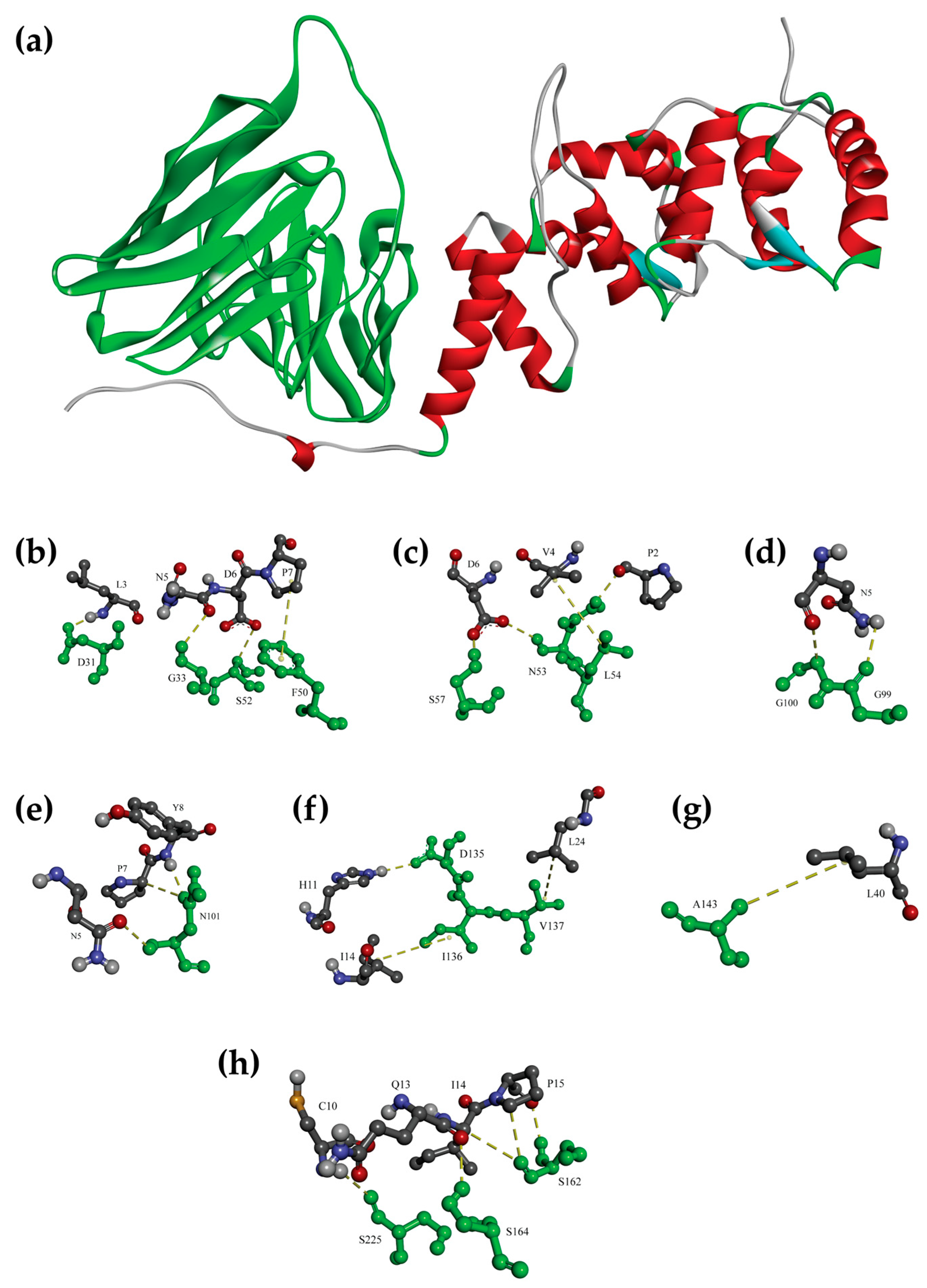
| OvROPN1L Amino Acids | scFv | Type of Interaction | |
|---|---|---|---|
| Amino Acids | Domain | ||
| * L3 | D31 | VH-CDR1 | H-bond |
| * N5 | G33 | VH-CDR1 | H-bond |
| * P7 | F50 | VH-FR2 | π-Alkyl |
| * D6 | S52 | VH-CDR2 | H-bond |
| * D6 | N53 | VH-CDR2 | H-bond |
| * P2 | H-bond | ||
| * V4 | L54 | VH-CDR2 | H-bond |
| * D6 | S57 | VH-CDR2 | H-bond |
| * N5 | G99 | VH-CDR3 | H-bond |
| * N5 | G100 | VH-CDR3 | H-bond |
| * N5 | N101 | VH-CDR3 | H-bond |
| * P7 | H-bond | ||
| * Y8 | H-bond | ||
| * H11 | D135 | VL-FR1 | H-bond |
| I14 | I136 | VL-FR1 | Hydrophobic |
| L24 | V137 | VL-FR1 | Hydrophobic |
| L40 | A143 | VL-FR1 | Hydrophobic |
| I14 | S162 | VL-CDR1 | H-bond |
| P15 | H-bond | ||
| * Q13 | S164 | VL-CDR1 | H-bond |
| * C10 | S225 | VL-CDR3 | H-bond |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geadkaew-Krenc, A.; Krenc, D.; Thanongsaksrikul, J.; Grams, R.; Phadungsil, W.; Glab-ampai, K.; Chantree, P.; Martviset, P. Production and Immunological Characterization of scFv Specific to Epitope of Opisthorchis viverrini Rhophilin-Associated Tail Protein 1-like (OvROPN1L). Trop. Med. Infect. Dis. 2023, 8, 160. https://doi.org/10.3390/tropicalmed8030160
Geadkaew-Krenc A, Krenc D, Thanongsaksrikul J, Grams R, Phadungsil W, Glab-ampai K, Chantree P, Martviset P. Production and Immunological Characterization of scFv Specific to Epitope of Opisthorchis viverrini Rhophilin-Associated Tail Protein 1-like (OvROPN1L). Tropical Medicine and Infectious Disease. 2023; 8(3):160. https://doi.org/10.3390/tropicalmed8030160
Chicago/Turabian StyleGeadkaew-Krenc, Amornrat, Dawid Krenc, Jeeraphong Thanongsaksrikul, Rudi Grams, Wansika Phadungsil, Kittirat Glab-ampai, Pathanin Chantree, and Pongsakorn Martviset. 2023. "Production and Immunological Characterization of scFv Specific to Epitope of Opisthorchis viverrini Rhophilin-Associated Tail Protein 1-like (OvROPN1L)" Tropical Medicine and Infectious Disease 8, no. 3: 160. https://doi.org/10.3390/tropicalmed8030160
APA StyleGeadkaew-Krenc, A., Krenc, D., Thanongsaksrikul, J., Grams, R., Phadungsil, W., Glab-ampai, K., Chantree, P., & Martviset, P. (2023). Production and Immunological Characterization of scFv Specific to Epitope of Opisthorchis viverrini Rhophilin-Associated Tail Protein 1-like (OvROPN1L). Tropical Medicine and Infectious Disease, 8(3), 160. https://doi.org/10.3390/tropicalmed8030160








