Abstract
The COVID-19 pandemic has disrupted the seasonal patterns of several infectious diseases. Understanding when and where an outbreak may occur is vital for public health planning and response. We usually rely on well-functioning surveillance systems to monitor epidemic outbreaks. However, not all countries have a well-functioning surveillance system in place, or at least not for the pathogen in question. We utilized Google Trends search results for RSV-related keywords to identify outbreaks. We evaluated the strength of the Pearson correlation coefficient between clinical surveillance data and online search data and applied the Moving Epidemic Method (MEM) to identify country-specific epidemic thresholds. Additionally, we established pseudo-RSV surveillance systems, enabling internal stakeholders to obtain insights on the speed and risk of any emerging RSV outbreaks in countries with imprecise disease surveillance systems but with Google Trends data. Strong correlations between RSV clinical surveillance data and Google Trends search results from several countries were observed. In monitoring an upcoming RSV outbreak with MEM, data collected from both systems yielded similar estimates of country-specific epidemic thresholds, starting time, and duration. We demonstrate in this study the potential of monitoring disease outbreaks in real time and complement classical disease surveillance systems by leveraging online search data.
1. Introduction
Respiratory syncytial virus (RSV) is the most common single cause of respiratory hospitalization of infants and is the second largest cause of lower respiratory infection mortality worldwide [1]. Currently there is no vaccine against RSV, although many preventive strategies are under development [2]. Historically, infection rates typically rise in late autumn and early winter in temperate climates. However, the seasonal patterns of several infectious diseases, including RSV, have been disrupted by the COVID-19 pandemic [3,4,5]. Specifically, RSV outbreaks were suppressed at the beginning of the COVID-19 pandemic (i.e., during the 2020–2021 period) [6] and resumed with irregular timing and increased magnitude from 2021–2022 onwards [7,8], e.g., see Japan [9], United States [10], United Kingdom [8], Turkey [11], Belgium [12] and Italy [13] in Northern Hemisphere, as well as Australia [14,15,16], New Zealand [17], Chile, Africa, Brazil [18] and other countries in the Southern Hemisphere [10]. These findings highlight the value of surveillance systems for RSV and other respiratory diseases during and after future pandemics, as the lifting of mitigation measures may result in severe outbreaks occurring with irregular timing [15]. Nevertheless, in most countries RSV is not a notifiable disease. Even in developed countries, surveillance systems have started monitoring RSV activity only recently. For example, New South Wales, Australia, only made RSV notifiable on 1 September 2022 [19].
Internet searches have previously been used to identify the timing, location, and magnitude of infectious disease outbreaks. In 2004, Johnson et al. reported a correlation between the frequency of influenza-related article access and the CDC’s surveillance data [20]. Similar results have been reported in Canada [21] and with Yahoo search queries [22]. In 2009, a study by Google and the CDC provided the first proof-of-concept for using Google search queries to detect influenza epidemics [23]. Since then, many influenza-related analyses have been conducted around the world, for example, in South Korea [24], Latin America [25], at a regional level in Manitoba, Canada [26] and South China, China [27], and at a city level in New York City [28] and Baltimore [29] in the US. Google Trends has also been used as a surveillance tool for other diseases, e.g., chicken pox [30], type 2 diabetes [31], dengue fever [32], Zika and Chikungunya [33], sexually transmitted infections [34], and COVID-19 [35].
Google Trends studies can be characterized into three main areas [36]: identifying seasonality, examining correlations between surveillance data and Google Trends, and forecasting. Seasonality has most commonly been investigated through visual observation [37,38,39,40,41,42], Kruskal–Wallis test [43,44,45], and Cosinor analysis [42,46,47,48,49]. The Pearson correlation coefficient is the principal statistical measure used to compare surveillance data to Google Trends [24,27,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64]. Forecasting is the least studied area. Only nine out of 104 studies reviewed in a 2018 systematic review were focused on prediction forecasting [36], with methods comprised of statistical modeling [30], linear regression [23,34,65,66], cross-correlation [58], and time series analysis (e.g., ARIMA [67]).
However, Google Trends as a surveillance tool has not been broadly applied to RSV, despite it being the leading cause of hospitalization in infants in developed countries. Two previous studies identified a correlation between Google Trends and RSV [68,69], while another found that searches for RSV can predict pediatric RSV encounters [70]. All of these studies have been retrospective. To our knowledge, there have been no studies focused on the prospective use of Google Trends as a surveillance tool to forecast the emergence of RSV outbreaks.
Our study aims to rigorously leverage real-time, online search engine data to nowcast emergence of RSV and consequently better manage the contemporary uncertainties enhanced by the ongoing COVID-19 pandemic. In this paper, we (1) investigated the correlation between RSV clinical surveillance data and Google Trends data observed at both country and city levels, (2) compared epidemiologic estimates derived from the Moving Epidemic Method (MEM) over Google Trends and clinical surveillance data, (3) discussed the use of Google Trends as a surveillance tool to nowcast the emergence of RSV outbreaks for countries where RSV surveillance data are limited.
2. Materials and Methods
2.1. Data
Google Trends data [71] reflect how a specific search interest varies for a region over time. It ranges from 100% to 0%, scaled by the highest search number that a specific search interest ever generated within the chosen time period. Weekly or monthly data points are shown if the chosen time period is shorter or longer than 5 years, accordingly. In this study, Japan, Germany, and Belgium were selected for illustration purposes because of their high quality surveillance data and Google Trends data. Five full years of weekly clinical and Google Trends search data were included for each country in this study. Among many options, one relevant keyword which shows the clearest seasonal pattern for each country was selected: “RS Virus” for Japan, “RS Virus” for Germany, and “RSV” for Belgium and the other 14 countries. Weekly RSV case data were gathered from each country’s official open access website, including the National Institute of Infectious Diseases [72] for Japan, the Robert Koch Institut [73] for Germany, and the Belgian Institute for Health [74] for Belgium.
2.2. Correlation
The Pearson correlation coefficient has been commonly used to assess the correlation between Google Trends search data and clinical surveillance data [64]. In this study, a Python library package, SciPy [75], was applied to perform the correlation analysis.
2.3. The Moving Epidemic Method (MEM)
The World Health Organization (WHO) released a guideline on assessing the severity of influenza in seasonal epidemics and pandemics in 2017 [76] and suggested two types of thresholds to characterize the start of an epidemic, one based on the Moving Epidemic Method (MEM) [77,78] and the other based on the pre-selected weekly positivity rates. However, using a certain percentage of RSV positivity among all RSV tests in a season as a threshold to define an epidemic [79,80,81] does not provide insights into the intensity of the epidemic and should not be used prospectively to detect the start of an epidemic [82]. In comparison, the epidemic threshold generated by MEM provides a good balance between sensitivity and specificity to detect seasonal epidemics and to avoid false alerts from data noises [77]. MEM has been widely adopted to calculate epidemic thresholds for both influenza [77,78,83,84] and RSV [82]. There have also been studies applying the MEM to multiple indicators (outpatient visits, hospitalization, and mortality rate) and combining multiple thresholds to classify the severity of an influenza season [85]. However, to our knowledge, MEM has not been applied over Google Trends data as an early-warning system for infectious disease outbreaks.
MEM is modeled based on historical data from a specific country or region. The method has three main steps [77,78]. In the first step, each season is separated into three periods: pre-epidemic, epidemic, and post-epidemic period. For each season separately, the length of the epidemic period is determined by an optimization process that maximizes positive cases within the least number of consecutive weeks. In the second step, the epidemic threshold is calculated as the upper limit of the 95% one-sided confidence interval of the arithmetic mean of the 30 highest pre-epidemic weekly rates from all seasons. The number of highest rates from each season is 30/(number of seasons). This epidemic threshold defines the start of the epidemic. In the third step, medium, high, and very high intensity thresholds are calculated as the upper limits of the 40%, 90%, and 97.5% one-sided confidence intervals of the geometric mean of 30 highest epidemic weekly rates. For the purpose of sensing the start of the epidemic, the first 2 steps are sufficient. The third step is to estimate the intensity of an epidemic. By dividing each season into three periods, the epidemic threshold is calculated only based on data points within the epidemic period, excluding false alerts of those abnormal high weekly rates during the pre-epidemic periods. By comparing the current week’s value against the epidemic threshold, we can know if the country being investigated is experiencing an epidemic period.
We first applied the MEM over countries with both clinical surveillance and Google Trends search data. The consistency of epidemiologic estimates derived from MEM with clinical surveillance vs. Google Trends search data were investigated to validate whether Google Trends can represent the clinical surveillance data in terms of estimating the epidemic starting week and duration. The ‘mem’ library in R was used in this study [86]. Google Trends search data applied with MEM were prepreprocessed with Loess transformation with default fixed criterium method provided in ‘R-mem’ library.
We also applied MEM to countries with limited publicly accessible clinical surveillance data. MEM thresholds estimated over the Google Trends data can be interpreted whether they are reliable or not without directly comparing to clinical case data. To interpret the reliability of the results, we can examine the goodness of fit of the MEM model using estimators such as sensitivity, specificity, positive predictive value, percent agreement, Matthews correlation coefficient, etc. In this paper, the Matthews correlation coefficient is reported. Details about how goodness of MEM is estimated are explained in [86]. Besides the goodness of fit, the epidemic percentage, which is the proportion of cases in the epidemic period over all cases, is generally a good indicator for understanding if there is a clear seasonality pattern and how well the MEM performed.
3. Results
3.1. Identical Seasonal Patterns between Google Trends and Case Data
Google Trends data matched case data with no delay in terms of seasonal start time, end time, and peak time for each epidemic in Japan and Germany (with Pearson correlation coefficient = 0.87, p-value < 0.0001 for Japan and Pearson correlation coefficient = 0.65, p-value < 0.0001 for Germany) (Figure 1). Note that weekly reports from Japan contained absolute case numbers from all sentinel hospitals, while reports from Germany contained positive test rates, where the test sample sizes ranged throughout the year. As a result, the correlation between case and search data for Germany was lower due to a higher fluctuation of cases. We did not apply any smoothing preprocessing on any of the data. These matched patterns were also found at the regional level (Figure 2). Tokyo and Kyoto analyses are presented here to illustrate this point.

Figure 1.
Correlation between Google Trends and case data at country level: Japan 2017–2021 (top) Pearson correlation coefficient = 0.87, p-value < 0.0001. Germany 2016–2020 (bottom) Pearson correlation coefficient = 0.65, p-value < 0.0001. Google Trends and case data are marked in blue and red accordingly. Y-axis for both Google Trends and case data ranges from the minimum to the maximum values of each function over time.
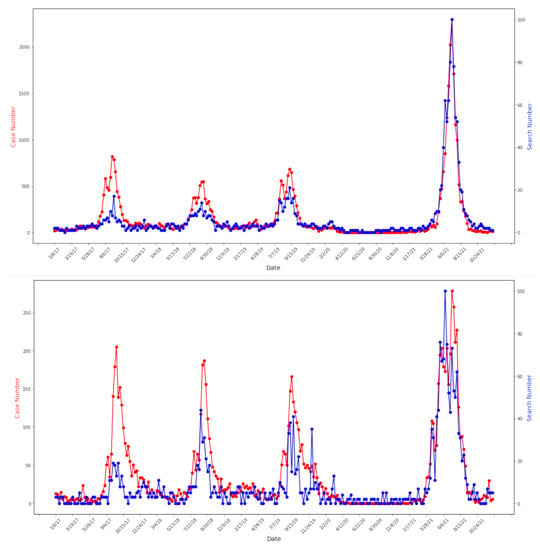
Figure 2.
Correlation between Google Trends and case data at regional level: Tokyo 2017–2021 (top) Pearson correlation coefficient = 0.92. Kyoto 2017–2021 (bottom) Pearson correlation coefficient = 0.83. Both p-value < 0.0001. Google Trends and case data are marked in blue and red accordingly. Y-axis for both Google Trends and case data ranges from the minimum to the maximum values of each function over time.
Google Trends data were also able to capture intra-annual abnormalities observed in case data. Using Belgium as an illustration, the seasonal patterns of RSV outbreaks can be seen in both case and search data shown in Figure 3. For the 2020–2021 season, the outbreak started later than previous years and had two peaks. This was also observed in the Google Trends data (shown above in Figure 3).

Figure 3.
RSV seasonality for Belgium: identical patterns within each season (i.e., bimodal for season 2020–2021, unimodal for other seasons) observed in both case (top) and Google Trends (bottom) data.
3.2. Identical Epidemiological Estimates from Case and Google Trends Data
Identical epidemic estimates in terms of season starting week and duration were obtained by applying MEM to Google Trends and clinical case data. In the cases of Japan and Germany, MEM provided identical estimates for the average start week and epidemic duration from the clinical case and Google Trends search data, with detailed results shown in Table 1.

Table 1.
Epidemic estimates of Japan 2015–2020 using MEM: identical epidemic seasonal start time and length were obtained by applying MEM over Google Trends and case data.
3.3. Epidemic Estimates from Google Trends Data in Countries with Limited Case Surveillance
We selected 14 countries without RSV clinical surveillance data to investigate using Google Trends. Using the same keyword “RSV”, seven out of 14 countries selected showed clear visual seasonal patterns (left panel of Figure 4) and the rest did not (right panel of Figure 4). Countries with clear visual seasonal patterns in Figure 4 generally correspond to higher epidemic percentage values. The purpose of using the same keyword is to illustrate how we can interpret whether the data with the chosen keyword is reliable or if the keyword is good enough to capture the pattern.
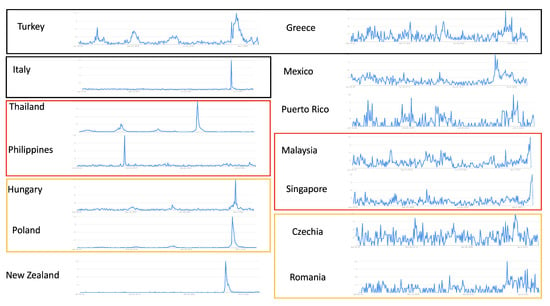
Figure 4.
Google Trends of countries with limited surveillance: using the same keyword “RSV”, countries with clear patterns are listed on the (left). Countries without clear patterns are listed on the (right). Countries that are close to each other geographically are boxed in the same color.
We then applied MEM to Google Trends data for the selected 14 countries to generate insightful epidemiologic estimates (Table 2). The surveillance column shows whether the countries are experiencing an epidemic based on MEM estimates. Current values for week 25, 2022 and thresholds were calculated based on the data of the five years before week 25, 2022, excluding 2020 and 2021 (due to abnormal RSV activities compared to other years) using MEM. A country is considered to be entering an epidemic if its current Google trend data value is above the estimated epidemic threshold dynamically generated by MEM. The column of 2020–2021 in Table 2 shows the start week and end week of last season. We are aware that the COVID-19 pandemic shifted the starting time for RSV outbreak in some countries. The 2020–2021 column is listed for a comparison to investigate whether this phenomenon can also be observed over the Google Trends data. Countries with clear visual seasonal patterns in terms of fewer fluctuations in Figure 4 on the left generally correspond to higher epidemic percentage values (Table 2 on the top), with a few countries as exceptions. As an unusually high RSV peak for Italy, the Philippines, Hungary, Thailand, Poland and New Zealand can dominate all other seasonal peaks and patterns in Google Trends, we attempted a few data selection heuristics to improve the method’s fitting results. One choice is to heuristically exclude abnormal, one-time-only epidemic peak data. For example, excluding Google Trends data for 2020 and 2021 for Hungary, Poland, Thailand and New Zealand resulted in an improved fitting performance (evaluated by the epidemic percentage value). By contrast, the low fitting performances for the Philippines and Italy have not yet been resolved, partially due to the facts that the abnormal peak is outside 2020 and 2021 and the identified Google Search data has an authentic unclear seasonal pattern, respectively.

Table 2.
MEM estimates for countries with limited surveillance for week 25, 2022: Estimates were based on Google Trends data between week 25, 2022 and previous 5 years, excluding 2020 and 2021 due to their abnormal intensified RSV activities. The upper part of the table contains countries listed on the left in Figure 4 with clear patterns. The lower part of the table contains countries listed on the right in Figure 4 with no clear patterns. Both parts are ranked by epidemic percentage, which is the ratio of cases during the epidemic to all cases.
4. Discussion
Google Trends can complement existing surveillance systems for monitoring disease outbreaks in real time. Using RSV as a case study, we revealed the strong correlation between Google Trends and clinical case data from Japan and Germany. We also observed that although many countries generate high quality case data, weekly reports may be delayed for several weeks due to various reasons. Google Trends can be used as a supplemental surveillance system for countries with limited sentinel network coverage, as well.
Google Trends is also not linked to the number of sentinel hospitals or the variation in reporting between testing sites. Most countries may not be able to extend their surveillance systems to collect data from all hospitals on time or maintain a reliable testing sample size across different times of the year. The positive testing rates may be sensitive to the testing sample size, creating false alarms as a result.
In our multiple country comparison effort, we also observed that Google Trends data were of a higher quality among countries with better surveillance systems. This may be due to socioeconomic factors such as better public health education that drove information seeking behavior online.
Occasionally, a single keyword such as “RSV” or “RS Virus” could be sufficient for identifying the clear seasonality patterns for RSV in Google Trends in certain countries, but not all: each country’s most suitable keyword for monitoring RSV outbreaks is still highly dependent on the local language choice. Unlike flu, adding additional keywords describing the disease symptoms may weaken the patterns, as many respiratory pathogens share a common pool of flu-like symptoms. Additionally, preprocessing the search data and then using MEM could prevent the false alarms caused by noisy fluctuation in the trends.
However, there can be issues with Google Trends. For example, Google Trends data are scaled based on the highest value in the time frame of choice. The abnormally high volume of Google Trends searches in 2020–2021 due to the COVID-19 pandemic scales down the rest of the normal seasons, which diminishes their seasonality patterns. The peak in 2020–2021 caused the epidemic threshold to considerably shift up compared to previous years. Therefore, when applying MEM to estimate country-specific outbreak thresholds, we excluded data from 2020 to 2021 because travel restrictions were in place in most countries. However, when obtaining data from Google Trends, 2020–2021 data were included to keep the current data point on the same scale as previous years, since Google Trends cannot exclude certain years. Additionally, monthly instead of weekly Google Trends data will be displayed if the time period selected on the platform is specified to be longer than five years. When there are no clinical case data to compare Google Trends data against, we can examine the epidemic percentage as an indicator of how the MEM performed. If the epidemic percentage is low, the estimates from MEM from Google Trends data may not be reliable. For example, although Italy had clear seasonal Google Trends patterns, searches at the start of the COVID-19 pandemic diminished compared to previous years, leaving the epidemic percentage value low and unreliable. One possible solution may be referring to the results from nearby countries in the same geographic region (Figure 4).
Notably, when the disease is not that well known (such as RSV), people tend to search for multiple keywords. Since respiratory diseases share similar symptoms, it may be challenging to collect accurate keywords for a particular disease or identify a seasonal pattern specific caused by a specific disease.
Additionally, both lower search volumes and clinical cases were observed after vaccination was introduced for other diseases such as rotavirus [87]. It remains unclear how much this would affect the correlation between clinical case and Google Trends search data for RSV, or if predicting using Google Trends data remains as sensitive as before a vaccine was introduced.
5. Conclusions
Google Trends can complement existing surveillance systems to monitor disease outbreaks in real time, especially in countries with limited or no sentinel network surveillance. Search data correlated well with clinical case data when both were available. Identical estimates of epidemic start time and duration were obtained from MEM using both Google Trends and clinical case data. The quality of clinical case data from countries with surveillance systems is linked to the sentinel hospital surveillance systems. This further identifies the importance of using alternative data streams, such as internet search data, to assist in locations where surveillance systems are not well established.
Author Contributions
D.W.: Conceptualization, Data Curation, Formal Analysis, Investigation, Methodology, Writing—Original Draft Preparation, Visualization, Writing—Review and Editing; A.G.: Conceptualization, Validation, Writing—Review and Editing; F.W.: Conceptualization, Supervision, Writing—Review and Editing; J.C.L.: Writing—Review and Editing; K.B.: Writing—Review and Editing; A.W.L.: Conceptualization, Supervision, Writing—Review and Editing; L.F.: Writing—Review and Editing; Y.-H.C.: Conceptualization, Project Administration, Supervision, Writing—Review and Editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank the cross-functional team in RSV clinical trial planning for their valuable comments.
Conflicts of Interest
All authors are the employees of Merck and MSD and may own stock and/or hold stock options in the company. This research collaboration was fully funded by the company.
Abbreviations
The following abbreviations are used in this manuscript:
| COVID-19 | Coronavirus disease |
| RSV | Respiratory syncytial virus |
| MEM | Moving Epidemic Method |
References
- Coultas, J.A.; Smyth, R.; Openshaw, P.J. Respiratory syncytial virus (RSV): A scourge from infancy to old age. Thorax 2019, 74, 986–993. [Google Scholar] [CrossRef] [PubMed]
- Battles, M.B.; McLellan, J.S. Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 2019, 17, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, L.; Sheppard, M.; Smith, A.; Dietz, S.; Jayanthi, P.; Yuan, Y.; Bull, L.; Wotiz, S.; Schwarze, T.; Azondekon, R.; et al. Changes in seasonal respiratory illnesses in the United States during the coronavirus disease 2019 (COVID-19) pandemic. Clin. Infect. Dis. 2021, 73, S110–S117. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H.; Song, K.H.; Kim, E.S.; Park, J.S.; Jung, J.; Ahn, S.; Jeong, E.K.; Park, H.; Kim, H.B. Impact of public health interventions on seasonal influenza activity during the COVID-19 outbreak in Korea. Clin. Infect. Dis. 2021, 73, e132–e140. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Xu, M.; Wang, X.; Xie, Y.; Du, X.; Chen, T.; Yang, L.; Wang, D.; Shu, Y. Nonpharmaceutical interventions used to control COVID-19 reduced seasonal influenza transmission in China. J. Infect. Dis. 2020, 222, 1780–1783. [Google Scholar] [CrossRef]
- Di Mattia, G.; Nenna, R.; Mancino, E.; Rizzo, V.; Pierangeli, A.; Villani, A.; Midulla, F. During the COVID-19 pandemic where has respiratory syncytial virus gone? Pediatr. Pulmonol. 2021, 56, 3106–3109. [Google Scholar] [CrossRef]
- Castagno, E.; Raffaldi, I.; Del Monte, F.; Garazzino, S.; Bondone, C. New epidemiological trends of respiratory syncytial virus bronchiolitis during COVID-19 pandemic. World J. Pediatr. 2022, 1–3. [Google Scholar] [CrossRef]
- Taylor, A.; Whittaker, E. The changing epidemiology of respiratory viruses in children during the COVID-19 pandemic: A Canary in a COVID Time. Pediatr. Infect. Dis. J. 2022, 41, e46. [Google Scholar] [CrossRef]
- Ohnishi, T.; Kawano, Y. Resurgence of respiratory syncytial virus infection during an atypical season in Japan. J. Pediatr. Infect. Dis. Soc. 2021, 10, 982–983. [Google Scholar] [CrossRef]
- Olsen, S.J.; Azziz-Baumgartner, E.; Budd, A.P.; Brammer, L.; Sullivan, S.; Pineda, R.F.; Cohen, C.; Fry, A.M. Decreased influenza activity during the COVID-19 pandemic—United States, Australia, Chile, and South Africa, 2020. Am. J. Transplant. 2020, 20, 3681–3685. [Google Scholar] [CrossRef]
- Agca, H.; Akalin, H.; Saglik, I.; Hacimustafaoglu, M.; Celebi, S.; Ener, B. Changing epidemiology of influenza and other respiratory viruses in the first year of COVID-19 pandemic. J. Infect. Public Health 2021, 14, 1186–1190. [Google Scholar] [CrossRef] [PubMed]
- Van Brusselen, D.; De Troeyer, K.; Ter Haar, E.; Vander Auwera, A.; Poschet, K.; Van Nuijs, S.; Bael, A.; Stobbelaar, K.; Verhulst, S.; Van Herendael, B.; et al. Bronchiolitis in COVID-19 times: A nearly absent disease? Eur. J. Pediatr. 2021, 180, 1969–1973. [Google Scholar] [CrossRef]
- Vittucci, A.C.; Piccioni, L.; Coltella, L.; Ciarlitto, C.; Antilici, L.; Bozzola, E.; Midulla, F.; Palma, P.; Perno, C.F.; Villani, A. The disappearance of respiratory viruses in children during the COVID-19 pandemic. Int. J. Environ. Res. Public Health 2021, 18, 9550. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, S.G.; Carlson, S.; Cheng, A.C.; Chilver, M.B.; Dwyer, D.E.; Irwin, M.; Kok, J.; Macartney, K.; MacLachlan, J.; Minney-Smith, C.; et al. Where has all the influenza gone? The impact of COVID-19 on the circulation of influenza and other respiratory viruses, Australia, March to September 2020. Eurosurveillance 2020, 25, 2001847. [Google Scholar] [CrossRef] [PubMed]
- Eden, J.S.; Sikazwe, C.; Xie, R.; Deng, Y.M.; Sullivan, S.G.; Michie, A.; Levy, A.; Cutmore, E.; Blyth, C.C.; Britton, P.N.; et al. Off-season RSV epidemics in Australia after easing of COVID-19 restrictions. Nat. Commun. 2022, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Saravanos, G.L.; Hu, N.; Homaira, N.; Muscatello, D.J.; Jaffe, A.; Bartlett, A.W.; Wood, N.J.; Rawlinson, W.; Kesson, A.; Lingam, R.; et al. RSV Epidemiology in Australia before and during COVID-19. Pediatrics 2022, 149. [Google Scholar] [CrossRef]
- Huang, Q.S.; Wood, T.; Jelley, L.; Jennings, T.; Jefferies, S.; Daniells, K.; Nesdale, A.; Dowell, T.; Turner, N.; Campbell-Stokes, P.; et al. Impact of the COVID-19 nonpharmaceutical interventions on influenza and other respiratory viral infections in New Zealand. Nat. Commun. 2021, 12, 1–7. [Google Scholar] [CrossRef]
- Varela, F.H.; Scotta, M.C.; Polese-Bonatto, M.; Sartor, I.T.S.; Ferreira, C.F.; Fernandes, I.R.; Zavaglia, G.O.; de Almeida, W.A.F.; Arakaki-Sanchez, D.; Pinto, L.A.; et al. Absence of detection of RSV and influenza during the COVID-19 pandemic in a Brazilian cohort: Likely role of lower transmission in the community. J. Glob. Health 2021, 11, 5007. [Google Scholar] [CrossRef]
- Goverment, N. Respiratory Syncytial Virus (RSV) Fact Sheet. Available online: https://www.health.nsw.gov.au/Infectious/factsheets/Pages/respiratory-syncytial-virus.aspx (accessed on 10 December 2022).
- Johnson, H.A.; Wagner, M.M.; Hogan, W.R.; Chapman, W.; Olszewski, R.T.; Dowling, J.; Barnas, G. Analysis of Web access logs for surveillance of influenza. In MEDINFO 2004; IOS Press: Amsterdam, The Netherlands, 2004; pp. 1202–1206. [Google Scholar]
- Eysenbach, G. Infodemiology: Tracking flu-related searches on the web for syndromic surveillance. In Proceedings of the AMIA Annual Symposium Proceedings, Washington, DC, USA, 11–15 November 2006; American Medical Informatics Association: Rockville, MD, USA, 2006; Volume 2006, p. 244. [Google Scholar]
- Polgreen, P.M.; Chen, Y.; Pennock, D.M.; Nelson, F.D.; Weinstein, R.A. Using internet searches for influenza surveillance. Clin. Infect. Dis. 2008, 47, 1443–1448. [Google Scholar] [CrossRef]
- Ginsberg, J.; Mohebbi, M.H.; Patel, R.S.; Brammer, L.; Smolinski, M.S.; Brilliant, L. Detecting influenza epidemics using search engine query data. Nature 2009, 457, 1012–1014. [Google Scholar] [CrossRef]
- Cho, S.; Sohn, C.H.; Jo, M.W.; Shin, S.Y.; Lee, J.H.; Ryoo, S.M.; Kim, W.Y.; Seo, D.W. Correlation between national influenza surveillance data and google trends in South Korea. PLoS ONE 2013, 8, e81422. [Google Scholar] [CrossRef] [PubMed]
- Pollett, S.; Boscardin, W.J.; Azziz-Baumgartner, E.; Tinoco, Y.O.; Soto, G.; Romero, C.; Kok, J.; Biggerstaff, M.; Viboud, C.; Rutherford, G.W. Evaluating Google Flu Trends in Latin America: Important lessons for the next phase of digital disease detection. Clin. Infect. Dis. 2016, 64, ciw657. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.T.; Gumel, A.; Thompson, L.H.; Strome, T.; Mahmud, S.M. “Google flu trends” and emergency department triage data predicted the 2009 pandemic H1N1 waves in Manitoba. Can. J. Public Health 2011, 102, 294–297. [Google Scholar] [CrossRef]
- Kang, M.; Zhong, H.; He, J.; Rutherford, S.; Yang, F. Using google trends for influenza surveillance in South China. PLoS ONE 2013, 8, e55205. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.R.; Konty, K.J.; Paladini, M.; Viboud, C.; Simonsen, L. Reassessing Google Flu Trends data for detection of seasonal and pandemic influenza: A comparative epidemiological study at three geographic scales. PLoS Comput. Biol. 2013, 9, e1003256. [Google Scholar] [CrossRef]
- Dugas, A.F.; Hsieh, Y.H.; Levin, S.R.; Pines, J.M.; Mareiniss, D.P.; Mohareb, A.; Gaydos, C.A.; Perl, T.M.; Rothman, R.E. Google Flu Trends: Correlation with emergency department influenza rates and crowding metrics. Clin. Infect. Dis. 2012, 54, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Bakker, K.M.; Martinez-Bakker, M.E.; Helm, B.; Stevenson, T.J. Digital epidemiology reveals global childhood disease seasonality and the effects of immunization. Proc. Natl. Acad. Sci. USA 2016, 113, 6689–6694. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, N.; Chotvijit, S.; Gupta, N.; Bradley, E.; Gilks, C.; Guo, W.; Crosby, H.; Shore, E.; Thiarai, M.; Procter, R.; et al. Google Trends can improve surveillance of Type 2 diabetes. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Husnayain, A.; Fuad, A.; Lazuardi, L. Correlation between Google Trends on dengue fever and national surveillance report in Indonesia. Glob. Health Action 2019, 12, 1552652. [Google Scholar] [CrossRef]
- Strauss, R.; Lorenz, E.; Kristensen, K.; Eibach, D.; Torres, J.; May, J.; Castro, J. Investigating the utility of Google trends for Zika and Chikungunya surveillance in Venezuela. BMC Public Health 2020, 20, 1–6. [Google Scholar] [CrossRef]
- Johnson, A.K.; Bhaumik, R.; Tabidze, I.; Mehta, S.D. Nowcasting sexually transmitted infections in Chicago: Predictive modeling and evaluation study using Google Trends. JMIR Public Health Surveill. 2020, 6, e20588. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.; Hopkins, C.; Surda, P. Use of Google Trends to investigate loss-of-smell–related searches during the COVID-19 outbreak. In International Forum of Allergy & Rhinology; Wiley Online Library: Hoboken, NJ, USA, 2020; Volume 10, pp. 839–847. [Google Scholar]
- Mavragani, A.; Ochoa, G.; Tsagarakis, K.P. Assessing the methods, tools, and statistical approaches in Google Trends research: Systematic review. J. Med. Internet Res. 2018, 20, e9366. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Harréus, U. Medical nowcasting using Google trends: Application in otolaryngology. Eur. Arch.-Oto-Rhino-Laryngol. 2013, 270, 2157–2160. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.G.; Song, W.J.; Choi, S.; Kim, H.; Ha, H.; Kim, S.H.; Cho, S.H.; Min, K.U.; Yoon, S.; Chang, Y.S. Google unveils a glimpse of allergic rhinitis in the real world. Allergy 2015, 70, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Seifter, A.; Schwarzwalder, A.; Geis, K.; Aucott, J. The utility of “Google Trends” for epidemiological research: Lyme disease as an example. Geospat. Health 2010, 4, 135–137. [Google Scholar] [CrossRef]
- Takada, K. Japanese interest in “Hotaru”(fireflies) and “Kabuto-Mushi”(japanese Rhinoceros beetles) corresponds with seasonality in visible abundance. Insects 2012, 3, 424–431. [Google Scholar] [CrossRef]
- Willson, T.J.; Lospinoso, J.; Weitzel, E.; McMains, K. Correlating regional aeroallergen effects on internet search activity. Otolaryngol.-Head Neck Surg. 2015, 152, 228–232. [Google Scholar] [CrossRef]
- Toosi, B.; Kalia, S. Seasonal and geographic patterns in tanning using real-time data from Google Trends. JAMA Dermatol. 2016, 152, 215–217. [Google Scholar] [CrossRef]
- Harsha, A.K.; Schmitt, J.E.; Stavropoulos, S.W. Know your market: Use of online query tools to quantify trends in patient information-seeking behavior for varicose vein treatment. J. Vasc. Interv. Radiol. 2014, 25, 53–57. [Google Scholar] [CrossRef]
- El-Sheikha, J. Global search demand for varicose vein information on the internet. Phlebology 2015, 30, 533–540. [Google Scholar] [CrossRef]
- Harsha, A.K.; Schmitt, J.E.; Stavropoulos, S.W. Match day: Online search trends reflect growing interest in IR training. J. Vasc. Interv. Radiol. 2015, 26, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Ingram, D.G.; Matthews, C.K.; Plante, D.T. Seasonal trends in sleep-disordered breathing: Evidence from Internet search engine query data. Sleep Breath. 2015, 19, 79–84. [Google Scholar] [CrossRef] [PubMed]
- DeVilbiss, E.A.; Lee, B.K. Brief report: Trends in US National autism awareness from 2004 to 2014: The impact of national autism awareness month. J. Autism Dev. Disord. 2014, 44, 3271–3273. [Google Scholar] [CrossRef] [PubMed]
- Plante, D.T.; Ingram, D.G. Seasonal trends in tinnitus symptomatology: Evidence from Internet search engine query data. Eur. Arch.-Oto-Rhino-Laryngol. 2015, 272, 2807–2813. [Google Scholar] [CrossRef]
- Sentana-Lledo, D.; Barbu, C.M.; Ngo, M.N.; Wu, Y.; Sethuraman, K.; Levy, M.Z. Seasons, searches, and intentions: What the internet can tell us about the bed bug (Hemiptera: Cimicidae) epidemic. J. Med. Entomol. 2016, 53, 116–121. [Google Scholar] [CrossRef]
- Zhang, Z.; Zheng, X.; Zeng, D.D.; Leischow, S.J. Tracking dabbing using search query surveillance: A case study in the United States. J. Med. Internet Res. 2016, 18, e5802. [Google Scholar] [CrossRef]
- Schootman, M.; Toor, A.; Cavazos-Rehg, P.; Jeffe, D.B.; McQueen, A.; Eberth, J.; Davidson, N.O. The utility of Google Trends data to examine interest in cancer screening. BMJ Open 2015, 5, e006678. [Google Scholar] [CrossRef]
- Bragazzi, N.L.; Dini, G.; Toletone, A.; Brigo, F.; Durando, P. Leveraging big data for exploring occupational diseases-related interest at the level of scientific community, media coverage and novel data streams: The example of silicosis as a pilot study. PLoS ONE 2016, 11, e0166051. [Google Scholar] [CrossRef]
- Bragazzi, N.L.; Watad, A.; Brigo, F.; Adawi, M.; Amital, H.; Shoenfeld, Y. Public health awareness of autoimmune diseases after the death of a celebrity. Clin. Rheumatol. 2017, 36, 1911–1917. [Google Scholar] [CrossRef]
- Gahr, M.; Uzelac, Z.; Zeiss, R.; Connemann, B.J.; Lang, D.; Schoenfeldt-Lecuona, C. Linking annual prescription volume of antidepressants to corresponding web search query data: A possible proxy for medical prescription behavior? J. Clin. Psychopharmacol. 2015, 35, 681–685. [Google Scholar] [CrossRef]
- Mavragani, A.; Sypsa, K.; Sampri, A.; Tsagarakis, K.P. Quantifying the UK online interest in substances of the EU watchlist for water monitoring: Diclofenac, estradiol, and the macrolide antibiotics. Water 2016, 8, 542. [Google Scholar] [CrossRef]
- Domnich, A.; Panatto, D.; Signori, A.; Lai, P.L.; Gasparini, R.; Amicizia, D. Age-related differences in the accuracy of web query-based predictions of influenza-like illness. PLoS ONE 2015, 10, e0127754. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.K.; Mehta, S.D. A comparison of Internet search trends and sexually transmitted infection rates using Google trends. Sex. Transm. Dis. 2014, 41, 61–63. [Google Scholar] [CrossRef]
- Wang, H.W.; Chen, D.R.; Yu, H.W.; Chen, Y.M. Forecasting the incidence of dementia and dementia-related outpatient visits with Google trends: Evidence from Taiwan. J. Med. Internet Res. 2015, 17, e264. [Google Scholar] [CrossRef]
- Crowson, M.G.; Schulz, K.; Tucci, D.L. National utilization and forecasting of ototopical antibiotics: Medicaid data versus “dr. google”. Otol. Neurotol. 2016, 37, 1049–1054. [Google Scholar] [CrossRef]
- Bragazzi, N.L.; Dini, G.; Toletone, A.; Brigo, F.; Durando, P. Infodemiological data concerning silicosis in the USA in the period 2004–2010 correlating with real-world statistical data. Data Brief 2017, 10, 457–464. [Google Scholar] [CrossRef]
- Bragazzi, N.L.; Bacigaluppi, S.; Robba, C.; Nardone, R.; Trinka, E.; Brigo, F. Infodemiology of status epilepticus: A systematic validation of the Google Trends-based search queries. Epilepsy Behav. 2016, 55, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Bragazzi, N.L. A Google Trends-based approach for monitoring NSSI. Psychol. Res. Behav. Manag. 2014, 7, 1. [Google Scholar] [CrossRef]
- Cavazos-Rehg, P.A.; Krauss, M.J.; Spitznagel, E.L.; Lowery, A.; Grucza, R.A.; Chaloupka, F.J.; Bierut, L.J. Monitoring of non-cigarette tobacco use using Google Trends. Tob. Control 2015, 24, 249–255. [Google Scholar] [CrossRef]
- Nuti, S.V.; Wayda, B.; Ranasinghe, I.; Wang, S.; Dreyer, R.P.; Chen, S.I.; Murugiah, K. The use of google trends in health care research: A systematic review. PLoS ONE 2014, 9, e109583. [Google Scholar] [CrossRef]
- Zhou, X.; Ye, J.; Feng, Y. Tuberculosis surveillance by analyzing Google trends. IEEE Trans. Biomed. Eng. 2011, 58, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Milinovich, G.J.; Avril, S.M.; Lê Cao, K.A.; Tong, S.; Hu, W. Disease surveillance based on Internet-based linear models: An Australian case study of previously unmodeled infection diseases. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Solano, P.; Ustulin, M.; Pizzorno, E.; Vichi, M.; Pompili, M.; Serafini, G.; Amore, M. A Google-based approach for monitoring suicide risk. Psychiatry Res. 2016, 246, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, H.A.; Mylonakis, E. Google trends: A web-based tool for real-time surveillance of disease outbreaks. Clin. Infect. Dis. 2009, 49, 1557–1564. [Google Scholar] [CrossRef] [PubMed]
- Uda, K.; Hagiya, H.; Yorifuji, T.; Koyama, T.; Tsuge, M.; Yashiro, M.; Tsukahara, H. Correlation Between National Surveillance and Search Engine Query Data on Respiratory Syncytial Virus Infections in Japan. Res. Sq. 2022. [Google Scholar] [CrossRef] [PubMed]
- Crowson, M.G.; Witsell, D.; Eskander, A. Using Google Trends to predict pediatric respiratory syncytial virus encounters at a major health care system. J. Med. Syst. 2020, 44, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Google Trends. Available online: https://trends.google.com/trends/ (accessed on 10 December 2022).
- NIID. IDWR Surveillance Data. Available online: https://www.niid.go.jp/niid/en/survaillance-data-table-english.html (accessed on 10 December 2022).
- Institute, R.K. Arbeitsgemeinschaft Influenza. Available online: https://influenza.rki.de (accessed on 10 December 2022).
- Sciensano. Weekly Flu Bulletin. Available online: https://www.sciensano.be/en/biblio/griep-bulletin-week-23-2022-bulletin-grippe-semaine-23-2022-weekly-flu-bulletin-week-23-2022 (accessed on 10 December 2022).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- World Health Organization. Pandemic Influenza Severity Assessment (PISA): A WHO Guide to Assess the Severity of Influenza in Seasonal Epidemics and Pandemics; Technical Report; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Vega, T.; Lozano, J.E.; Meerhoff, T.; Snacken, R.; Mott, J.; Ortiz de Lejarazu, R.; Nunes, B. Influenza surveillance in Europe: Establishing epidemic thresholds by the moving epidemic method. Influenza Other Respir. Viruses 2013, 7, 546–558. [Google Scholar] [CrossRef]
- Vega, T.; Lozano, J.E.; Meerhoff, T.; Snacken, R.; Beauté, J.; Jorgensen, P.; Ortiz de Lejarazu, R.; Domegan, L.; Mossong, J.; Nielsen, J.; et al. Influenza surveillance in E urope: Comparing intensity levels calculated using the moving epidemic method. Influenza Other Respir. Viruses 2015, 9, 234–246. [Google Scholar] [CrossRef]
- Broberg, E.K.; Waris, M.; Johansen, K.; Snacken, R.; Penttinen, P. Seasonality and geographical spread of respiratory syncytial virus epidemics in 15 European countries, 2010 to 2016. Eurosurveillance 2018, 23, 17-00284. [Google Scholar] [CrossRef]
- Bloom-Feshbach, K.; Alonso, W.J.; Charu, V.; Tamerius, J.; Simonsen, L.; Miller, M.A.; Viboud, C. Latitudinal variations in seasonal activity of influenza and respiratory syncytial virus (RSV): A global comparative review. PLoS ONE 2013, 8, e54445. [Google Scholar] [CrossRef] [PubMed]
- Obando-Pacheco, P.; Justicia-Grande, A.J.; Rivero-Calle, I.; Rodríguez-Tenreiro, C.; Sly, P.; Ramilo, O.; Mejías, A.; Baraldi, E.; Papadopoulos, N.G.; Nair, H.; et al. Respiratory syncytial virus seasonality: A global overview. J. Infect. Dis. 2018, 217, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Vos, L.M.; Teirlinck, A.C.; Lozano, J.E.; Vega, T.; Donker, G.A.; Hoepelman, A.I.; Bont, L.J.; Oosterheert, J.J.; Meijer, A. Use of the moving epidemic method (MEM) to assess national surveillance data for respiratory syncytial virus (RSV) in the Netherlands, 2005 to 2017. Eurosurveillance 2019, 24, 1800469. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.L.; Marques, D.F.; Cameron, R.L.; Potts, A.; Bishop, J.; von Wissmann, B.; William, N.; Reynolds, A.J.; Robertson, C.; McMenamin, J. Moving epidemic method (MEM) applied to virology data as a novel real time tool to predict peak in seasonal influenza healthcare utilisation. The Scottish experience of the 2017/18 season to date. Eurosurveillance 2018, 23, 18–00079. [Google Scholar] [CrossRef] [PubMed]
- Rakocevic, B.; Grgurevic, A.; Trajkovic, G.; Mugosa, B.; Grujicic, S.S.; Medenica, S.; Bojovic, O.; Alonso, J.E.L.; Vega, T. Influenza surveillance: Determining the epidemic threshold for influenza by using the Moving Epidemic Method (MEM), Montenegro, 2010/11 to 2017/18 influenza seasons. Eurosurveillance 2019, 24, 1800042. [Google Scholar] [CrossRef]
- Biggerstaff, M.; Kniss, K.; Jernigan, D.B.; Brammer, L.; Bresee, J.; Garg, S.; Burns, E.; Reed, C. Systematic assessment of multiple routine and near real-time indicators to classify the severity of influenza seasons and pandemics in the United States, 2003–2004 through 2015–2016. Am. J. Epidemiol. 2018, 187, 1040–1050. [Google Scholar] [CrossRef]
- Lozano, J.E. The Moving Epidemic Method Shiny Web Application. Available online: https://github.com/lozalojo/memapp (accessed on 10 December 2022).
- Burnett, E.; Parashar, U.D.; Winn, A.; Tate, J.E. Trends in Rotavirus Laboratory Detections and Internet Search Volume Before and After Rotavirus Vaccine Introduction and in the Context of the Coronavirus Disease 2019 Pandemic—United States, 2000–2021. J. Infect. Dis. 2022, 226, 967–974. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).