The Distribution of Eight Antimicrobial Resistance Genes in Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii Strains Isolated from Dental Plaque as Oral Commensals
Abstract
1. Introduction
2. Materials and Methods
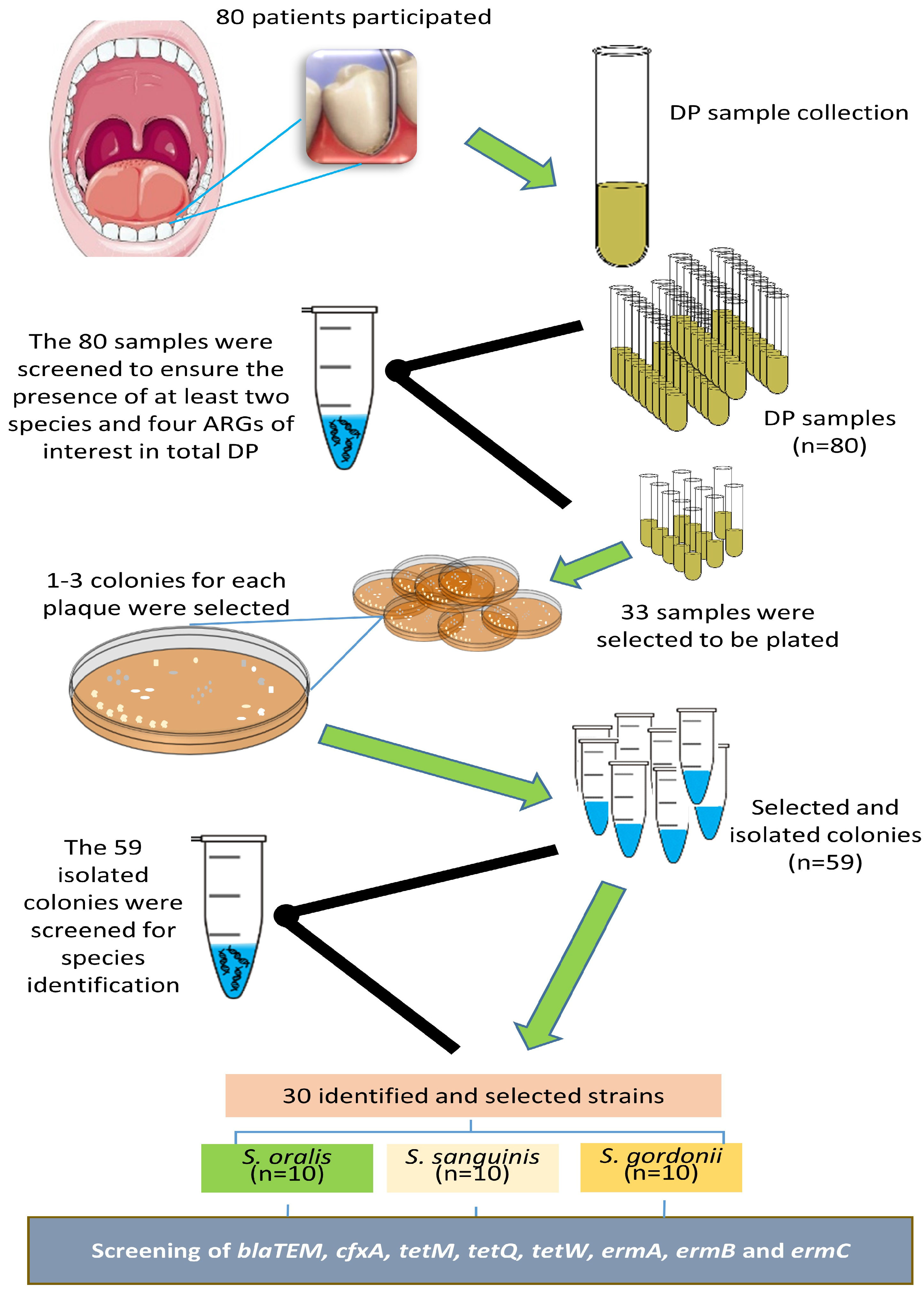
2.1. Study Population and Clinical Evaluation
2.2. Sample Collection
2.3. Bacteria and ARG Identification in the DP Samples
2.4. Culture and Identification of Streptococci from Selected DP Samples
2.5. Identification of ARGs in the Selected Isolated Streptococci
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, M.; Umashankar, D.N.; Viswanath, D.; Girish, G. Role of the Oral Microflora in Health and Disease. J. Indian Acad. Oral Med. Radiol. 2013, 25, 184–187. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Curtis, H.; Blaser, M.J.; Dirk, G.; Kota, K.C.; Rob, K.; Liu, B.; Wang, L.; Sahar, A.; White, J.R.; Badger, J.H. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral Microbiomes: More and More Importance in Oral Cavity and Whole Body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.A.; Diene, S.M.; Rolain, J.M. Human Microbiomes and Antibiotic Resistance. Hum. Microbiome J. 2018, 10, 43–52. [Google Scholar] [CrossRef]
- Costalonga, M.; Herzberg, M.C. The Oral Microbiome and the Immunobiology of Periodontal Disease and Caries. Immunol. Lett. 2014, 162, 22–38. [Google Scholar] [CrossRef]
- Baty, J.J.; Stoner, S.N.; Scoffield, J.A. Oral Commensal Streptococci: Gatekeepers of the Oral Cavity. J. Bacteriol. 2022, 204, e00257-22. [Google Scholar] [CrossRef] [PubMed]
- Bik, E.M.; Long, C.D.; Armitage, G.C.; Loomer, P.; Emerson, J.; Mongodin, E.F.; Nelson, K.E.; Gill, S.R.; Fraser-Liggett, C.M.; Relman, D.A. Bacterial Diversity in the Oral Cavity of 10 Healthy Individuals. ISME J. 2010, 4, 962–974. [Google Scholar] [CrossRef]
- Tagg, J.R.; Wescombe, P.A.; Hale, J.D.F.; Burton, J.P. Streptococcus: A Brief Update on the Current Taxonomic Status of the Genus. In Lactic Acid Bacteria; CRC Press: Boca Raton, FL, USA, 2019; pp. 87–107. [Google Scholar]
- Okahashi, N.; Nakata, M.; Kuwata, H.; Kawabata, S. Oral Mitis Group Streptococci: A Silent Majority in Our Oral Cavity. Microbiol. Immunol. 2022, 66, 539–551. [Google Scholar] [CrossRef]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef]
- Marsh, P.D. Dental Plaque: Biological Significance of a Biofilm and Community Life-style. J. Clin. Periodontol. 2005, 32, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.; Mullany, P. Oral Biofilms: A Reservoir of Transferable, Bacterial, Antimicrobial Resistance. Expert Rev. Anti. Infect. Ther. 2010, 8, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Arif, B.; Caetano-Anollés, G.; Kim, K.M.; Nasir, A. Horizontal Gene Transfer in Human-Associated Microorganisms Inferred by Phylogenetic Reconstruction and Reconciliation. Sci. Rep. 2019, 9, 5953. [Google Scholar] [CrossRef] [PubMed]
- Johnsborg, O.; Eldholm, V.; Håvarstein, L.S. Natural Genetic Transformation: Prevalence, Mechanisms and Function. Res. Microbiol. 2007, 158, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Cvitkovitch, D.G. Genetic Competence and Transformation in Oral Streptococci. Crit. Rev. Oral Biol. Med. 2001, 12, 217–243. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, L.; Wahl, A.; Fléchard, M.; Mignolet, J.; Hols, P. Regulation of Competence for Natural Transformation in Streptococci. Infect. Genet. Evol. 2015, 33, 343–360. [Google Scholar] [CrossRef] [PubMed]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic Resistance Genes in Bacteria: Occurrence, Spread, and Control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria—A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Meek, R.W.; Vyas, H.; Piddock, L.J.V. Nonmedical Uses of Antibiotics: Time to Restrict Their Use? PLoS Biol. 2015, 13, e1002266. [Google Scholar] [CrossRef]
- Nadeem, S.F.; Gohar, U.F.; Tahir, S.F.; Mukhtar, H.; Pornpukdeewattana, S.; Nukthamna, P.; Moula Ali, A.M.; Bavisetty, S.C.B.; Massa, S. Antimicrobial Resistance: More than 70 Years of War between Humans and Bacteria. Crit. Rev. Microbiol. 2020, 46, 578–599. [Google Scholar] [CrossRef]
- Moraes, L.C.; Só, M.V.R.; Da Silva Dal Pizzol, T.; Ferreira, M.B.C.; Montagner, F. Distribution of Genes Related to Antimicrobial Resistance in Different Oral Environments: A Systematic Review. J. Endod. 2015, 41, 434–441. [Google Scholar] [CrossRef]
- Brooks, L.; Narvekar, U.; McDonald, A.; Mullany, P. Prevalence of Antibiotic Resistance Genes in the Oral Cavity and Mobile Genetic Elements That Disseminate Antimicrobial Resistance: A Systematic Review. Mol. Oral Microbiol. 2022, 37, 133–153. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.O.; Gabany, J.; Persson, G.R.; Roberts, M.C. Distribution of Erm (F) and Tet (Q) Genes in 4 Oral Bacterial Species and Genotypic Variation between Resistant and Susceptible Isolates. J. Clin. Periodontol. 2002, 29, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Takano, K.; Maeda, N. Distribution of the Tetracycline Resistance Determinant TetQ Gene in Oral Isolates of Black-pigmented Anaerobes in Japan. Oral Microbiol. Immunol. 2001, 16, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Ramos, V.R.; Pérez-Serrano, R.M.; García-Solís, P.; Solís-Sainz, J.C.; Espinosa-Cristóbal, L.F.; Castro-Ruíz, J.E.; Domínguez-Pérez, R.A. Root Canal Microbiota as an Augmented Reservoir of Antimicrobial Resistance Genes in Type 2 Diabetes Mellitus Patients Abstract. J. Appl. Oral Sci. 2023, 30, e20220362. [Google Scholar] [CrossRef] [PubMed]
- Abe, F.C.; Kodaira, K.; Motta, C.D.C.B.; Barberato-Filho, S.; Silva, M.T.; Guimarães, C.C.; Martins, C.C.; Lopes, L.C. Antimicrobial Resistance of Microorganisms Present in Periodontal Diseases: A Systematic Review and Meta-Analysis. Front. Microbiol. 2022, 13, 961986. [Google Scholar] [CrossRef] [PubMed]
- Rôças, I.N.; Siqueira, J.F., Jr. Detection of Antibiotic Resistance Genes in Samples from Acute and Chronic Endodontic Infections and after Treatment. Arch. Oral Biol. 2013, 58, 1123–1128. [Google Scholar] [CrossRef]
- Sukumar, S.; Roberts, A.P.; Martin, F.E.; Adler, C.J. Metagenomic Insights into Transferable Antibiotic Resistance in Oral Bacteria. J. Dent. Res. 2016, 95, 969–976. [Google Scholar] [CrossRef]
- Mansfield, J.M.; Herrmann, P.; Jesionowski, A.M.; Vickerman, M.M. Streptococcus Gordonii Pheromone Sg CAM373 May Influence the Reservoir of Antibiotic Resistance Determinants of Enterococcus Faecalis Origin in the Oral Metagenome. J. Med. Microbiol. 2017, 66, 1635. [Google Scholar] [CrossRef]
- Herrero, E.R.; Slomka, V.; Bernaerts, K.; Boon, N.; Hernandez-Sanabria, E.; Passoni, B.B.; Quirynen, M.; Teughels, W. Antimicrobial Effects of Commensal Oral Species Are Regulated by Environmental Factors. J. Dent. 2016, 47, 23–33. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the Wild: Antibiotic Resistance Genes in Natural Environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Sommer, M.O.A.; Munck, C.; Toft-Kehler, R.V.; Andersson, D.I. Prediction of Antibiotic Resistance: Time for a New Preclinical Paradigm? Nat. Rev. Microbiol. 2017, 15, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Loyola-Rodriguez, J.P.; Galvan-Torres, L.J.; Martinez-Martinez, R.E.; Abud-Mendoza, C.; Medina-Solis, C.E.; Ramos-Coronel, S.; Garcia-Cortes, J.O.; Domínguez-Pérez, R.A. Frequency of Dental Caries in Active and Inactive Systemic Lupus Erythematous Patients: Salivary and Bacterial Factors. Lupus 2016, 25, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Armitage, G.C. Development of a Classification System for Periodontal Diseases and Conditions. Ann. Periodontol. 1999, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, T.; Kawaguchi, M.; Shimizu, N.; Hoshino, N.; Ooshima, T.; Fujiwara, T. PCR Detection and Identification of Oral Streptococci in Saliva Samples Using Gtf Genes. Diagn. Microbiol. Infect. Dis. 2004, 48, 195–199. [Google Scholar] [CrossRef]
- Rôças, I.N.; Siqueira, J.F., Jr. Antibiotic Resistance Genes in Anaerobic Bacteria Isolated from Primary Dental Root Canal Infections. Anaerobe 2012, 18, 576–580. [Google Scholar] [CrossRef]
- Lancaster, H.; Bedi, R.; Wilson, M.; Mullany, P. The Maintenance in the Oral Cavity of Children of Tetracycline-Resistant Bacteria and the Genes Encoding Such Resistance. J. Antimicrob. Chemother. 2005, 56, 524–531. [Google Scholar] [CrossRef]
- Ready, D.; Pratten, J.; Roberts, A.P.; Bedi, R.; Mullany, P.; Wilson, M. Potential Role of Veillonella spp. as a Reservoir of Transferable Tetracycline Resistance in the Oral Cavity. Antimicrob. Agents Chemother. 2006, 50, 2866–2868. [Google Scholar] [CrossRef]
- Milanović, V.; Aquilanti, L.; Tavoletti, S.; Garofalo, C.; Osimani, A.; De Filippis, F.; Ercolini, D.; Ferrocino, I.; Cagno, R.D.; Turroni, S. Distribution of Antibiotic Resistance Genes in the Saliva of Healthy Omnivores, Ovo-Lacto-Vegetarians, and Vegans. Genes 2020, 11, 1088. [Google Scholar] [CrossRef]
- King, A.; Bathgate, T.; Phillips, I. Erythromycin Susceptibility of Viridans Streptococci from the Normal Throat Flora of Patients Treated with Azithromycin or Clarithromycin. Clin. Microbiol. Infect. 2002, 8, 85–92. [Google Scholar] [CrossRef][Green Version]
- Villedieu, A.; Diaz-Torres, M.L.; Roberts, A.P.; Hunt, N.; McNab, R.; Spratt, D.A.; Wilson, M.; Mullany, P. Genetic Basis of Erythromycin Resistance in Oral Bacteria. Antimicrob. Agents Chemother. 2004, 48, 2298–2301. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, I.; Sakellari, D.; Spala, A.; Arsenakis, M.; Konstantinidis, A. Prevalence of TetM, TetQ, Nim and BlaTEM Genes in the Oral Cavities of Greek Subjects: A Pilot Study. J. Clin. Periodontol. 2009, 36, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Talebi Bezmin Abadi, A.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization Report: Current Crisis of Antibiotic Resistance. Bionanoscience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- Binta, B.; Patel, M. Detection of CfxA2, CfxA3, and CfxA6 Genes in Beta-Lactamase Producing Oral Anaerobes. J. Appl. Oral Sci. 2016, 24, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Sedgley, C.M.; Lee, E.H.; Martin, M.J.; Flannagan, S.E. Antibiotic Resistance Gene Transfer between Streptococcus Gordonii and Enterococcus Faecalis in Root Canals of Teeth Ex Vivo. J. Endod. 2008, 34, 570–574. [Google Scholar] [CrossRef]
- Bryskier, A. Viridans Group Streptococci: A Reservoir of Resistant Bacteria in Oral Cavities. Clin. Microbiol. Infect. 2002, 8, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Luna, V.A.; Coates, P.; Eady, E.A.; Cove, J.H.; Nguyen, T.T.H.; Roberts, M.C. A Variety of Gram-Positive Bacteria Carry Mobile Mef Genes. J. Antimicrob. Chemother. 1999, 44, 19–25. [Google Scholar] [CrossRef]
- Kataja, J.; Huovinen, P.; Skurnik, M.; Finnish Study Group for Antimicrobial Resistance; Seppälä, H. Erythromycin Resistance Genes in Group A Streptococci in Finland. Antimicrob. Agents Chemother. 1999, 43, 48–52. [Google Scholar] [CrossRef]
- Yumoto, H.; Hirota, K.; Hirao, K.; Ninomiya, M.; Murakami, K.; Fujii, H.; Miyake, Y. The Pathogenic Factors from Oral Streptococci for Systemic Diseases. Int. J. Mol. Sci. 2019, 20, 4571. [Google Scholar] [CrossRef]
- Lucas, V.S.; Gafan, G.; Dewhurst, S.; Roberts, G.J. Prevalence, Intensity and Nature of Bacteraemia after Toothbrushing. J. Dent. 2008, 36, 481–487. [Google Scholar] [CrossRef]
- Lockhart, P.B.; Brennan, M.T.; Thornhill, M.; Michalowicz, B.S.; Noll, J.; Bahrani-Mougeot, F.K.; Sasser, H.C. Poor Oral Hygiene as a Risk Factor for Infective Endocarditis–Related Bacteremia. J. Am. Dent. Assoc. 2009, 140, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.P.; Warner, M.; Broughton, K.; James, D.; Efsratiou, A.; George, R.C.; Livermore, D.M. Antibiotic Susceptibility of Streptococci and Related Genera Causing Endocarditis: Analysis of UK Reference Laboratory Referrals, January 1996 to March 2000. BMJ 2001, 322, 395–396. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shenep, J.L. Viridans-Group Streptococcal Infections in Immunocompromised Hosts. Int. J. Antimicrob. Agents. 2000, 14, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Luby, E.; Ibekwe, A.M.; Zilles, J.; Pruden, A. Molecular Methods for Assessment of Antibiotic Resistance in Agricultural Ecosystems: Prospects and Challenges. J. Environ. Qual. 2016, 45, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, V.; Day, M.R.; Doumith, M.; Hopkins, K.L.; Woodford, N.; Godbole, G.; Jenkins, C. Comparison of Phenotypic and WGS-Derived Antimicrobial Resistance Profiles of Enteroaggregative Escherichia coli Isolated from Cases of Diarrhoeal Disease in England, 2015–2016. J. Antimicrob. Chemother. 2017, 72, 3288–3297. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Serrano, R.M.; Domínguez-Pérez, R.A.; Ayala-Herrera, J.L.; Luna-Jaramillo, A.E.; de Larrea, G.Z.-L.; Solís-Sainz, J.C.; García-Solís, P.; Loyola-Rodríguez, J.P. Dental Plaque Microbiota of Pet Owners and Their Dogs as a Shared Source and Reservoir of Antimicrobial Resistance Genes. J. Glob. Antimicrob. Resist. 2020, 21, 285–290. [Google Scholar] [CrossRef]
| Streptococci Species | Primer Sequence (5′-3′) | Annealing Temp (°C) | Amplicon Size (bp) |
|---|---|---|---|
| S. sanguinis | GGATAGTGGCTCAGGGCAGCCAGTT GAACAGTTGCTGGACTTGCTTCTC | 70 | 313 |
| S. oralis | TCCCGGTCAGCAAACTCCAGCC GCAACCTTTGGGATTTGCAAC | 66 | 374 |
| S. gordonii | CTATGCGGATGATGCTAATCAAGTG GGAGYCGCTATAATCTTGTCAGAAA | 55 | 440 |
| ARGs | Primer Sequence (5′-3′) | Annealing Temp (°C) | Amplicon Size (bp) |
| blaTEM | CCAATGCTTAATCAGTGAGG ATGAGTATTCAACATTTCCG | 50 | 858 |
| cfxA | GCGCAAATCCTCCTTTAACAA ACCGCCACACCAATTTCG | 55 | 802 |
| tetM | GTGGACAAAGGTAC AACGAG CGGTAAAGTTCGTCACACAC | 55 | 406 |
| tetW | GAGAGCCTGCTATATGCCAGC GGGCGTATCCACAATGTTAAC | 55 | 168 |
| tetQ | TTATACTTCCTCCGGC ATCG ATCGGTTCGAGAATGTCCAC | 55 | 904 |
| ermA | AACACCCTGAACCCAAGGGACG CTTCACATCCGGATTCGCTCGA | 50 | 420 |
| ermB | GAAAAGGTACTCAACCAAATA AGTAACGGTACTTAAATTGTTTAC | 55 | 639 |
| ermC | AATC GGCTCAGGAAAAGG ATCGTCAATTCCTGCATG | 50 | 562 |
| Number of Species | Frequency (%) |
|---|---|
| Three | 36 (45) |
| Two | 25 (31.2) |
| One | 16 (20) |
| None | 3 (3.7) |
| Number of ARGs | Frequency (%) |
|---|---|
| Eight | 0 |
| Seven | 0 |
| Six | 6 (7.5) |
| Five | 20 (25) |
| Four | 13 (16.2) |
| Three | 11 (13.7) |
| Two | 12 (15) |
| One | 9 (11.2) |
| None | 9 (11.2) |
| Specific ARG | Frequency (%) |
|---|---|
| tetM | 69 (86.2) |
| ermB | 46 (57.5) |
| blaTEM | 39 (48.7) |
| tetW | 33 (41.2) |
| ermC | 24 (30) |
| tetQ | 19 (23.7) |
| cfxA | 11 (13.7) |
| ermA | 8 (10) |
| S. oralis | 10 |
| S. sanguinis | 11 |
| S. gordonii | 16 |
| Others | 22 |
| S. sanguinis (n = 10) | S. oralis (n = 10) | S. gordonii (n = 10) | All Selected (n = 30) | |
|---|---|---|---|---|
| Specific ARG | Frequency (%) | |||
| tetM | 10 (100) | 6 (60) | 6 (60) | 22 (73.3) |
| blaTEM | 8 (80) | 8 (80) | 4 (40) | 20 (66.6) |
| tetW | 6 (60) | 8 (80) | 6 (60) | 20 (66.6) |
| ermC | 3 (30) | 6 (60) | 2 (20) | 11 (36.6) |
| ermB | 3 (30) | 2 (20) | 2 (20) | 7 (23.3) |
| tetQ | 2 (20) | 0 | 2 (20) | 4 (13.3) |
| ermA | 0 | 0 | 0 | 0 |
| cfxA | 0 | 0 | 0 | 0 |
| Number of Detected ARGs | Frequency (%) | |||
|---|---|---|---|---|
| Six | 1 (10) | 0 | 1 (10) | 2 (6.6) |
| Five | 0 | 0 | 0 | 0 |
| Four | 1 (10) | 6 (60) | 1 (10) | 8 (24.2) |
| Three | 6 (60) | 1 (10) | 1 (10) | 8 (24.2) |
| Two | 2 (20) | 1 (10) | 2 (20) | 5 (16.6) |
| One | 0 | 1 (10) | 2 (20) | 3 (10) |
| None | 0 | 1 (10) | 2 (20) | 3 (10) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Dorantes, V.; Domínguez-Pérez, R.A.; Pérez-Serrano, R.M.; Solís-Sainz, J.C.; García-Solís, P.; Espinosa-Cristóbal, L.F.; Cabeza-Cabrera, C.V.; Ayala-Herrera, J.L. The Distribution of Eight Antimicrobial Resistance Genes in Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii Strains Isolated from Dental Plaque as Oral Commensals. Trop. Med. Infect. Dis. 2023, 8, 499. https://doi.org/10.3390/tropicalmed8110499
Morales-Dorantes V, Domínguez-Pérez RA, Pérez-Serrano RM, Solís-Sainz JC, García-Solís P, Espinosa-Cristóbal LF, Cabeza-Cabrera CV, Ayala-Herrera JL. The Distribution of Eight Antimicrobial Resistance Genes in Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii Strains Isolated from Dental Plaque as Oral Commensals. Tropical Medicine and Infectious Disease. 2023; 8(11):499. https://doi.org/10.3390/tropicalmed8110499
Chicago/Turabian StyleMorales-Dorantes, Verónica, Rubén Abraham Domínguez-Pérez, Rosa Martha Pérez-Serrano, Juan Carlos Solís-Sainz, Pablo García-Solís, León Francisco Espinosa-Cristóbal, Claudia Verónica Cabeza-Cabrera, and José Luis Ayala-Herrera. 2023. "The Distribution of Eight Antimicrobial Resistance Genes in Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii Strains Isolated from Dental Plaque as Oral Commensals" Tropical Medicine and Infectious Disease 8, no. 11: 499. https://doi.org/10.3390/tropicalmed8110499
APA StyleMorales-Dorantes, V., Domínguez-Pérez, R. A., Pérez-Serrano, R. M., Solís-Sainz, J. C., García-Solís, P., Espinosa-Cristóbal, L. F., Cabeza-Cabrera, C. V., & Ayala-Herrera, J. L. (2023). The Distribution of Eight Antimicrobial Resistance Genes in Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii Strains Isolated from Dental Plaque as Oral Commensals. Tropical Medicine and Infectious Disease, 8(11), 499. https://doi.org/10.3390/tropicalmed8110499






