Genomic Analysis of Multidrug-Resistant Escherichia coli Strains Isolated in Tamaulipas, Mexico
Abstract
:1. Introduction
2. Materials and Methods
2.1. E. coli Strains
2.2. Isolation of Strains
2.3. Antimicrobial Susceptibility Testing
2.4. Whole Genome Sequencing
2.5. Bioinformatic Analysis
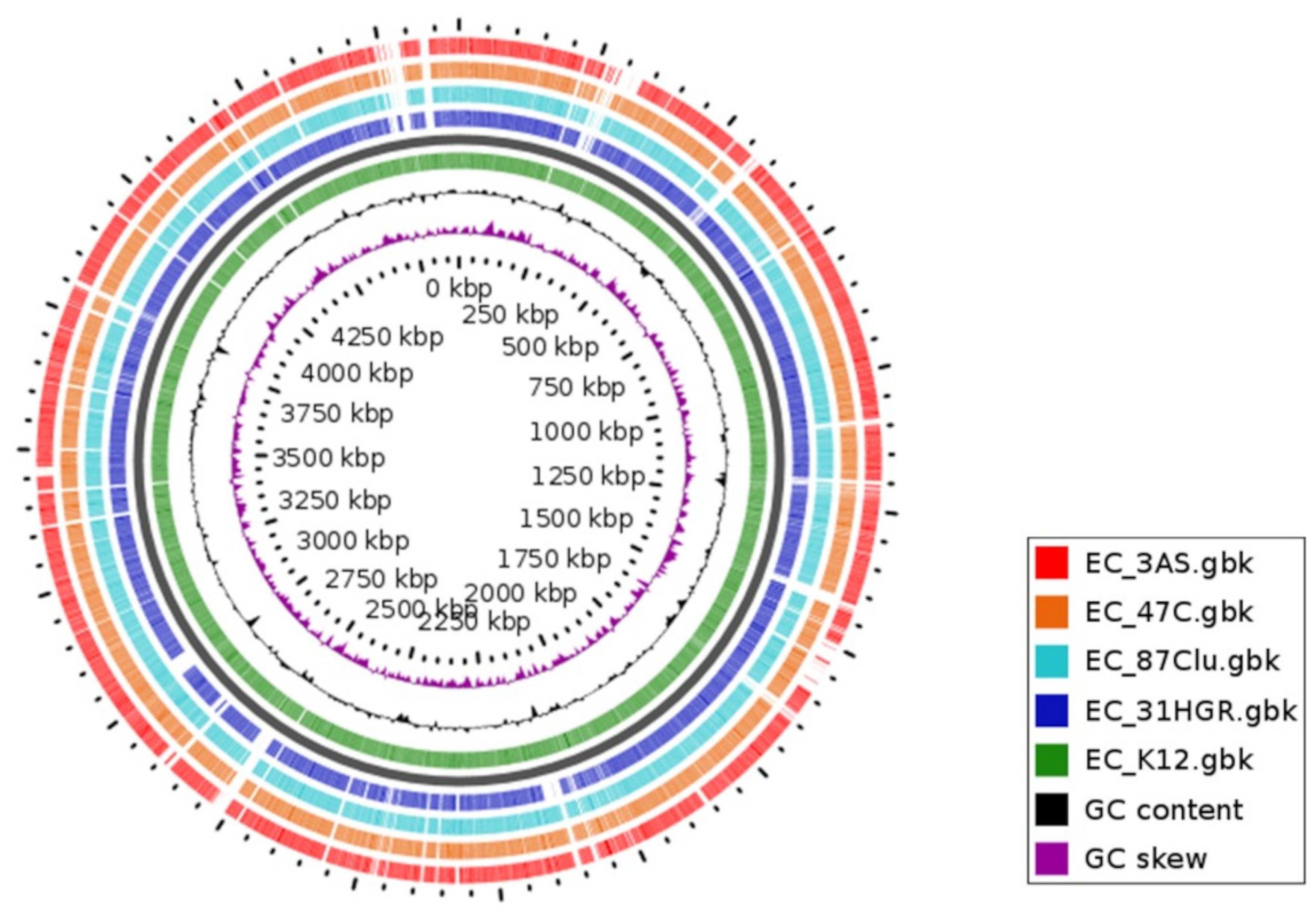
2.6. Phylogenomic Analysis and Genomic Comparison
3. Results
3.1. Antibiotic Susceptibility
3.2. Genomic Characteristics
3.3. Resistome
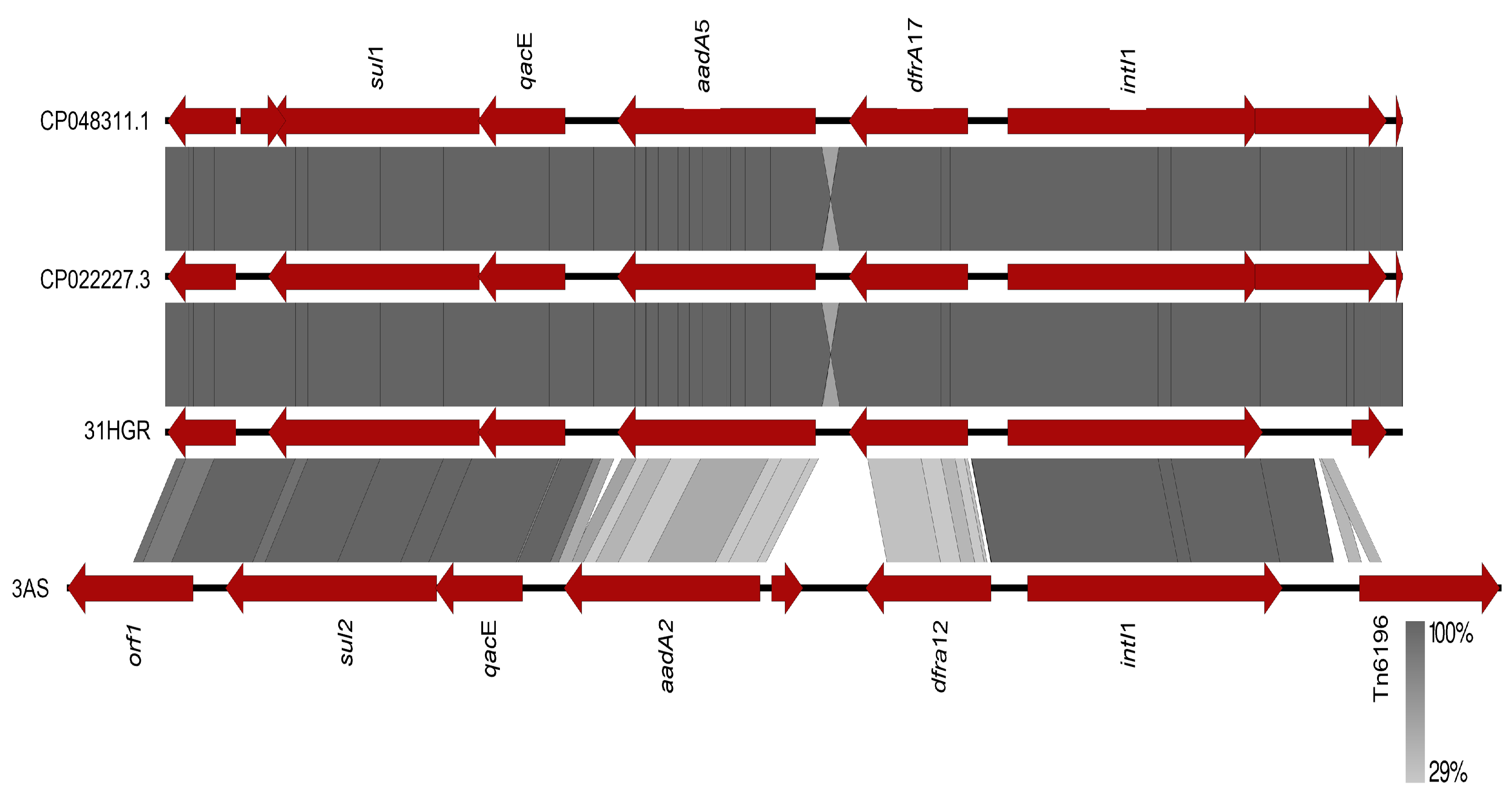
3.4. Mobilome of E. coli
3.4.1. Plasmids
3.4.2. Phages
3.4.3. Other EGMs
3.5. Virulome
3.6. Phylogenomic Analysis
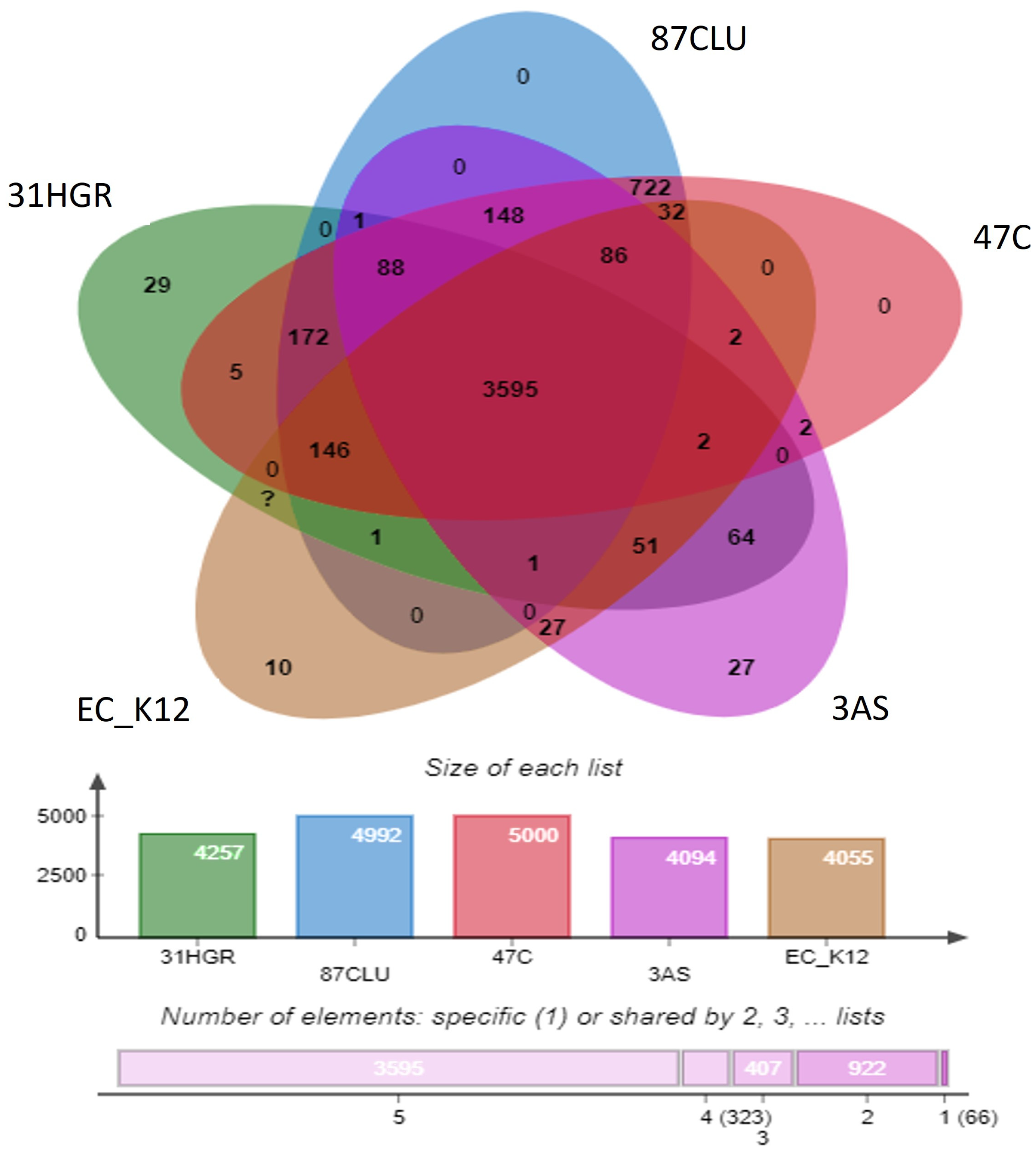
3.7. Comparative Genomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Rev. Antimicrob. Resist. 2016. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 13 September 2023).
- Aworh, M.K.; Kwaga, J.K.P.; Hendriksen, R.S.; Okolocha, E.C.; Thakur, S. Genetic Relatedness of Multidrug Resistant Escherichia coli Isolated from Humans, Chickens and Poultry Environments. Antimicrob. Resist. Infect. Control 2021, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- van Elsas, J.D.; Semenov, A.V.; Costa, R.; Trevors, J.T. Elsas Survival of Escherichia coli in the Environment: Fundamental and Public Health Aspects. ISME J. 2011, 5, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Mbelle, N.M.; Feldman, C.; Sekyere, J.O.; Maningi, N.E.; Modipane, L.; Essack, S.Y. The Resistome, Mobilome, Virulome and Phylogenomics of Multidrug-Resistant Escherichia coli Clinical Isolates from Pretoria, South Africa. Sci. Rep. 2019, 9, 16457. [Google Scholar] [CrossRef] [PubMed]
- Massella, E.; Giacometti, F.; Bonilauri, P.; Reid, C.J.; Djordjevic, S.P.; Merialdi, G.; Bacci, C.; Fiorentini, L.; Massi, P.; Bardasi, L.; et al. Antimicrobial Resistance Profile and ExPEC Virulence Potential in Commensal Escherichia coli of Multiple Sources. Antibiotics 2021, 10, 351. [Google Scholar] [CrossRef]
- Shin, S.W.; Shin, K.; Jung, M.; Belaynehe, M.; Yoo, S. Prevalence of Antimicrobial Resistance and Transfer of Tetracycline Resistance Genes in Escherichia coli Isolates from Beef Cattle. Appl. Environ. Microbiol. 2015, 81, 5560–5566. [Google Scholar] [CrossRef]
- Reid, C.J.; Blau, K.; Jechalke, S.; Smalla, K.; Djordjevic, S.P.; Campo, R. Del Whole Genome Sequencing of Escherichia coli From Store-Bought Produce. Front. Microbiol. 2020, 10, 3050. [Google Scholar] [CrossRef]
- Reid, C.J.; Wyrsch, E.R.; Chowdhury, P.R.; Zingali, T.; Liu, M.; Darling, A.E.; Chapman, T.A.; Djordjevic, S.P. Porcine Commensal Escherichia coli: A Reservoir for Class 1 Integrons Associated with IS 26. Microb. Genom. 2017, 3, e000143. [Google Scholar] [CrossRef]
- Martínez-Vázquez, A.V.; Rivera-Sánchez, G.; Lira-Méndez, K.; Reyes-López, M.A.; Bocanegra-García, V. Prevalence, Antimicrobial Resistance and Virulence Genes of Escherichia coli Isolated from Retail Meat in Tamaulipas, Mexico. J. Glob. Antimicrob. Resist. 2018, 14, 266–272. [Google Scholar] [CrossRef]
- Martínez-Vázquez, A.V.; Vázquez-Villanueva, J.; Leyva-Zapata, L.M.; Barrios-García, H.; Rivera, G.; Bocanegra-García, V. Multidrug Resistance of Escherichia coli Strains Isolated From Bovine Feces and Carcasses in Northeast Mexico. Front. Vet. Sci. 2021, 8, 643802. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Vázquez, A.V.; Mandujano, A.; Cruz-Gonzalez, E.; Guerrero, A.; Vazquez, J.; Cruz-Pulido, W.L.; Rivera, G.; Bocanegra-García, V. Evaluation of Retail Meat as a Source of ESBL Escherichia coli. Antibiotics 2022, 11, 1795. [Google Scholar] [CrossRef] [PubMed]
- Moghnia, O.H.; Al-Sweih, N.A. Whole Genome Sequence Analysis of Multidrug Resistant Escherichia coli and Klebsiella pneumoniae Strains in Kuwait. Microorganisms 2022, 10, 507. [Google Scholar] [CrossRef]
- CLSI. M100 Performance Standards for Antimicrobial; CLSI: Malvern, PA, USA, 2018; ISBN 156238838X. [Google Scholar]
- Ortega-Balleza, J.L.; Guerrero, A.; Castro-Escarpulli, G.; Cruz-González, E.; Rivera, G.; Bocanegra-García, V. Draft Genome Sequence of a Uropathogenic Escherichia coli. Microbiol. Resour. Annouc. 2022, 11, e0093121. [Google Scholar]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding Data and Analysis Capabilities. Nucleic Acids Res. 2020, 48, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, 16–21. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A Web Server for Whole-Genome Comparison and Annotation of Orthologous Clusters across Multiple Species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Kot, B. Antibiotic Resistance Among Uropathogenic Escherichia coli. Pol. J. Microbiol. 2019, 68, 403–415. [Google Scholar] [CrossRef]
- Ramírez-Castillo; Castillo, F.Y.R.; Flores, A.C.M.; González, F.J.A.; Díaz, F.M.; Harel, J.; Barrera, A.L.G. An Evaluation of Multidrug—Resistant Escherichia coli Isolates in Urinary Tract Infections from Aguascalientes, Mexico: Cross—Sectional Study. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 34. [Google Scholar] [CrossRef] [PubMed]
- Gomi, R.; Matsuda, T.; Matsumura, Y.; Yamamoto, M.; Tanaka, M.; Ichiyama, S.; Yoneda, M. Crossm Resistant and Extraintestinal Pathogenic Escherichia coli in River Water. Appl. Environ. Microbiol. 2017, 83, e02703-16. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, N.; Aganj, M.; Ali, L.; Shokoohizadeh, L.; Sakinc, T. Frequency Distribution of Genes Encoding Aminoglycoside Modifying Enzymes in Uropathogenic E. coli Isolated from Iranian Hospital. BMC Res. Nores 2014, 7, 842. [Google Scholar] [CrossRef]
- Rocha-Gracia, R.C.; Lozano-Zarain, P.; Cázarez, Z.G.; Andrea, C.; Brambila, E.; Torres, C.; Cortés-Cortés, G. IncFIB Plasmids Carrying the Resistance Gene Bla CTX-M-15 in ESBL-Producing Escherichia coli Clones from Pediatric Patients. J. Infect. Dev. Ctries. 2022, 16, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, R.; Abdullah, A.; Ahmed, D.; Hussain, A. High Prevalence of blaCTX-M-15 Gene among Escherichia coli Isolates Causing Extraintestinal Infections in Bangladesh. Antibiotics 2020, 9, 796. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Kader, F.; Hamza, E.; Abdel-Moein, K.A.; Sabry, M.A. Retail Chicken Giblets Contaminated with Extended-Spectrum Cephalosporin-and Carbapenem-Resistant Salmonella enterica Carrying blaCMY-2. Vet. World 2022, 15, 1297–1304. [Google Scholar] [CrossRef]
- Jouini, A.; Klibi, A.; Kmiha, S.; Hamrouni, S.; Ghram, A.; Maaroufi, A. Lineages, Virulence Gene Associated and Integrons among Extended Spectrum β-Lactamase (ESBL) and CMY-2 Producing Enterobacteriaceae from Bovine Mastitis, in Tunisia. Pathogens 2022, 11, 948. [Google Scholar] [CrossRef]
- Fang, L.X.; Li, X.P.; Li, L.; Chen, M.Y.; Wu, C.Y.; Li, L.L.; Liao, X.-P.; Liu, Y.-H.; Sun, J. ISEcp1 -Mediated Transposition of Chromosome- Borne BlaCMY-2 into an Endogenous ColE1-like Plasmid in Escherichia coli. Infect. Drug Resist. 2018, 11, 995–1005. [Google Scholar] [CrossRef]
- Awosile, B.; Reyes-velez, J.; Cuesta-Astroz, Y.; Rodríguez-Lecompte, J.C.; Saab, M.E.; Heider, L.C.; Keefe, G.; Sánchez, J.; Mcclure, J.T. Whole-Genome Sequence Analysis of 4 Fecal blaCMY-2 -Producing Escherichia coli Isolates from Holstein Dairy Calves. J. Dairy Sci. 2020, 103, 877–883. [Google Scholar] [CrossRef]
- Merida-Vieyra, J.; De Colsa-Ranero, A.; Calderón-Castañeda, Y.; Aquino-Andrade, A. Detection of CMY-Type Beta-Lactamases in Escherichia coli Isolates from Paediatric Patients in a Tertiary Care Hospital in Mexico. Antimicrob. Resist. Infect. Control 2020, 9, 168. [Google Scholar] [CrossRef]
- Gomes, C.; Ruiz-Roldán, L.; Mateu, J.; Ochoa, T.J.; Ruiz, J. Azithromycin Resistance Levels and Mechanisms in Escherichia coli. Sci. Rep. 2019, 9, 6089. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.C.P.; Woerther, P.L.; Bouvet, M.; Andremont, A.; Leclercq, R.; Canu, A. Escherichia coli as Reservoir for Macrolide. Emerg. Infect. Dis. 2009, 15, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Wu, F.; Chai, Y.; Xu, X.; Yang, L.; Tian, S.; Zhang, H.; Li, Y.; Yang, C.; Liu, H.; et al. A New Plasmid Carrying MphA Causes Prevalence of Azithromycin Resistance in Enterotoxigenic Escherichia coli Serogroup. BMC Microbiol. 2020, 20, 247. [Google Scholar] [CrossRef] [PubMed]
- Reza, H.; Wang, P.; Robertson, I.; Abraham, S.; Sahibzada, S.; Habib, I. Antimicrobial Resistance and Genomic Characterisation of Escherichia coli Isolated from Caged and Non-Caged Retail Table Eggs in Western Australia. Int. J. Food Microbiol. 2021, 340, 109054. [Google Scholar] [CrossRef]
- Adesoji, A.; Ogunjobi, A.; Olatoye, I.; Douglas, D. Prevalence of Tetracycline Resistance Genes among Multi-Drug Resistant Bacteria from Selected Water Distribution Systems in Southwestern Nigeria. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 35. [Google Scholar] [CrossRef]
- Awad, A.; Arafat, N.; Elhadidy, M. Genetic Elements Associated with Antimicrobial Resistance among Avian Pathogenic Escherichia coli. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 59. [Google Scholar] [CrossRef]
- Ochoa, S.A.; Cruz-Cordova, A.; Lune-Pineda, V.M.; Reyes-Grajeda, J.P.; Cázares-Domínguez, V.; Escalona, G.; Sépulbeda-González, M.E.; López-Montiel, F.; Arellano-Galinedo, J.; López-Martínez, B.; et al. Multidrug- and Extensively Drug-Resistant Uropathogenic Escherichia coli Clinical Strains: Phylogenetic Groups Widely Associated with Integrons Maintain High Genetic Diversity. Front. Microbiol. 2016, 7, 2042. [Google Scholar] [CrossRef]
- Delgado-Blas, J.F.; Ovejero, C.M.; David, S.; Montero, N.; Calero-Caceres, W.; Garcillan-Barcia, M.P.; de la Cruz, F.; Muniesa, M.; Aanensen, D.M.; Gonzalez-Zorn, B. Population Genomics and Antimicrobial Resistance Dynamics of Escherichia coli in Wastewater and River Environments. Commun. Biol. 2021, 4, 457. [Google Scholar] [CrossRef]
- Neumann, B.; Rackwitz, W.; Hunfeld, K.P.; Fuchs, S.; Werner, G.; Pfeifer, Y. Genome Sequences of Two Clinical Escherichia coli Isolates Harboring the Novel. Gut Pathog. 2020, 12, 40. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, W.; Wang, H.; Zhao, S.; Chen, Y.; Meng, F.; Zhang, Y.; Xu, H.; Chen, X.; Zhang, F. Specific Patterns of GyrA Mutations Determine the Resistance Difference to Ciprofloxacin and Levofloxacin in Klebsiella pneumoniae and Escherichia coli. BMC Infect. Dis. 2013, 13, 2–7. [Google Scholar] [CrossRef]
- Astorga, F.; Navarrete-Talloni, M.J.; Miró, M.P.; Bravo, V.; Toro, M.; Blondel, C.J.; Hervé-Claude, L.P. Antimicrobial Resistance in E. coli Isolated from Dairy Calves and Bedding Material. Heliyon 2019, 5, e02773. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Chen, L. The Significance of Epidemic Plasmids in the Success of Multidrug-Resistant Drug Pandemic Extraintestinal Pathogenic Escherichia coli. Infect. Dis. Ther. 2023, 12, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids Carrying Antimicrobial Resistance Genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Crossm Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Mbanga, J.; Amoako, D.G.; Abia, A.L.K.; Allam, M. Genomic Insights of Multidrug-Resistant Escherichia coli From Wastewater Sources and Their Association With Clinical Pathogens in South Africa. Front. Vet. Sci. 2021, 8, 636715. [Google Scholar] [CrossRef] [PubMed]
- Furlan, R.; Lopes, R.; Santana, M.; David, L.; Rosa, S.; Angelino, E.; Stehling, G. Infection, Genetics and Evolution blaCTX-M-2 and blaCMY-2 Recovered from an Urban Stream. Infect. Genet. Evol. 2021, 96, 105156. [Google Scholar] [CrossRef]
- Garza-Montúfar, M.E.; Treviño-Valdez, P.D.; De La Garza-Salina, L. Comorbidities and Antimicrobial Resistance in Urological Outpatients with Positive Urine Culture. Rev. Med. Inst. Mex. Seguro Soc. 2018, 56, 347–353. [Google Scholar]
- Ballesteros-Monrreal, M.G.; Arenas-Hernández, M.M.P.; Enciso-Martínez, Y.; Martínez-de la Peña, C.F.; Rocha-Gracia, R.C.; Lozano-Zaraín, P.; Navarro-Ocaña, A.; Martínez-Laguna, Y. Virulence and Resistance Determinants of Uropathogenic Escherichia coli Strains Isolated from Pregnant and Non-Pregnant Women from Two States in Mexico. Infect. Drug Resist. 2020, 13, 295–310. [Google Scholar] [CrossRef]
- Paniagua-Contreras, G.L.; Monroy-Pérez, E.; Díaz-Velásquez, C.E.; Uribe-García, A.; Labastida, A.; Peñaloza-Figueroa, F.; Domínguez-Trejo, P.; García, L.R.; Vaca-Paniagua, F.; Vaca, S. Whole-Genome Sequence Analysis of Multidrug-Resistant Uropathogenic Strains of Escherichia coli from Mexico. Infect. Drug Resist. 2019, 12, 2363–2377. [Google Scholar] [CrossRef]
- Zhang, X.; Xiao, S.; Jiang, X.; Li, Y.; Fan, Z.; Yu, Y.; Wang, P.; Li, D. Genomic Characterization of Escherichia coli LCT—EC001, an Extremely Multidrug—Resistant Strain with an Amazing Number of Resistance Genes. Gut Pathog. 2019, 11, 25. [Google Scholar] [CrossRef]

| Isolate ID | City | Source | Year |
|---|---|---|---|
| 31HGR | Reynosa | Urine (human) | 2018 |
| 87CLU | Reynosa | Urine (human) | 2018 |
| 47C | Rio Bravo | Chicken retail meat | 2017 |
| 3AS | Diaz Ordaz | Surface water (Rio Grande River) | 2018 |
| Antimicrobial Family | Strains | |||
|---|---|---|---|---|
| 31HGR | 87CLU | 47C | 3AS | |
| Tetracyclines | TET MIN DOX | TET | TET MIN DOX | TET DOX |
| Penicillins | AMP | AMP | AMC AMP | AMC |
| Aminoglycosides | - | STR GE | STR GE | STR GE |
| Folate pathway antagonists | SXT | SXT | - | SXT |
| Quinolones | LEV CIP | LEV NA | CIP | - |
| Phenicols | - | CHL | CHL | CHL |
| Cephalosporins | CTX FEP | CTX CRO | CF | CF |
| Nitrofuran | - | - | NF | - |
| MDR | + | + | + | + |
| Features | 31HGR | 87CLU | 47C | 3AS |
|---|---|---|---|---|
| Source | Urine | Urine | Meat | Water |
| Contigs | 141 | 162 | 148 | 96 |
| N50 | 91,860 | 206,030 | 205,584 | 175,845 |
| Size (Mb) | 4.98 | 5.31 | 5.31 | 4.72 |
| GC content (%) | 50.82 | 50.80 | 50.82 | 50.95 |
| CDS | 4969 | 4690 | 5406 | 4690 |
| tRNA | 73 | 78 | 83 | 78 |
| rRNA | 6 | 10 | 9 | 10 |
| CRISPR repetition | 22 | 13 | 44 | 13 |
| CRISPR spacer | 20 | 12 | 42 | 12 |
| CRISPR arrays | 2 | 1 | 2 | 1 |
| Serotype | O101:H4 | O10:H23 | O10:H23 | O21:H21 |
| Sequence type (ST) | 44 | 224 | 224 | 155 |
| Isolate | SRA Number | Phenotypic Resistance Profile | ARG | MLST | Virulence Genes | Plasmid Replicon |
|---|---|---|---|---|---|---|
| 31HGR | SRR15258840 | SXT, CTX, LEV, FEP, AMP, CIP, TET, MIN, DOX | tet(B), blaOXA-1’ blaCTX-M-15’ aadA5 aph(6)-Id, aph(3″)-Ib, aac(6’)-Ib-cr, sul1, sul2, dfra17, catB3, qacE, mdfA, mph(A) | 44 | iucA, iutA, sitA, irp1, irp2, fyuA, ybt, csg, fimH, ompA, traT, hlyE | IncFIA, IncFIB, IncFII, IncY |
| 87CLU | SRR25437123 | CHL, SXT, CTX, LEV, AMP, STR, GE, NA, CRO, TET | tet(A), blaTEM-1B, blaCMY-2, aadA1, aadA2, cmlA1, aac3-IIa, aph(6)-Id, aph(3″)-Ia, aph(3″)-Ib, sul2, sul3, dfra14, floR, qacL, mdf(A) | 224 | iucA, iutA, sitA, hra, csg, fimH, IpfA, ompA, cvaC, gad, espP, cma, traT, hlyE | IncFIA, IncFIB, IncFII, IncY |
| 47C | SRR25602185 | STR, CF, GE, AMP, CHL, NF, AMC, CIP, TET, MIN, DOX | tet(A), tet(B), blaTEM1B, blaCMY2, aadA1, aadA2, cmlA1, aac3-IIa, aph(6)-Id, aph(3″)-Ia, aph(3″)-Ib, sul2, sul3, dfra14, floR, qacL, mdfA | 224 | iucA, iutA, sitA, hra, csg, fimH, IpfA, ompA, cvaC, gad, espP, cma, traT, hlyE | IncFIA, IncFIB, IncFII, IncY |
| 3AS | SRR25645458 | CF, GE, SXT, CHL, AMC, STR, TET, DOX | tet(A), aadA2, sul1, dfra12, floR, qnrB, qacE, mdfA | 155 | csg, fimH, IpfA, ompA, gad, hlyE | IncFIB, IncY, IncR, Col |
| Isolate ID | Mutations | Plasmid-Mediated | ||
|---|---|---|---|---|
| gyrA | parE | parC | qnrB | |
| 31HGR | S83L, D87N | S458T | S80I | ND |
| 87CLU | S83L, D87N | S458A | S80I | ND |
| 47C | S83L, D87N | S458A | S80I | ND |
| 3AS | ND | ND | ND | Detected |
| Strain | Intact Prophage | Region Length | GC % | Total Proteins | Most Common Phage |
|---|---|---|---|---|---|
| 31HGR | 1 | 43 | 49.85 | 56 | PHAGE_Escher_phiV10_NC_007804 |
| 2 | 35.1 | 51.84 | 33 | PHAGE_Escher_pro483_NC_028943 | |
| 3 | 45.3 | 50.15 | 53 | PHAGE_Entero_mEp460_NC_019716 | |
| 6 | 33.5 | 52.45 | 36 | PHAGE_Entero_lambda_NC_001416 | |
| 87CLU | 2 | 41.7 | 50.93 | 62 | PHAGE_Entero_SfV_NC_003444 |
| 4 | 31.7 | 50.67 | 32 | PHAGE_Klebsi_4LV2017_NC_047818 | |
| 11 | 13.6 | 55.02 | 20 | PHAGE_Entero_P88_NC_026014 | |
| 12 | 12.1 | 55.44 | 19 | PHAGE_Entero_P88_NC_026014 | |
| 13 | 9.2 | 53.04 | 16 | PHAGE_Escher_pro147_NC_028896 | |
| 47C | 1 | 41.7 | 50.93 | 61 | PHAGE_Entero_SfV_NC_003444 |
| 4 | 34.7 | 50.61 | 37 | PHAGE_Klebsi_4LV2017_NC_047818 | |
| 7 | 39.3 | 49.74 | 38 | PHAGE_Escher_pro147_NC_028896 | |
| 8 | 95.5 | 47.64 | 106 | PHAGE_Salmon_SJ46_NC_031129 | |
| 9 | 31.2 | 51.27 | 32 | PHAGE_Entero_fiAA91_ss_NC_022750 | |
| 10 | 12.2 | 55.43 | 19 | PHAGE_Entero_P88_NC_026014 | |
| 11 | 11.6 | 55.86 | 18 | PHAGE_Entero_P88_NC_026014 | |
| 3AS | 1 | 57.3 | 51.10 | 86 | PHAGE_Erwini_vB_EhrS_59_NC_048198 |
| 2 | 43.1 | 51.85 | 52 | PHAGE_Shigel_SfII_NC_021857 | |
| 3 | 23.8 | 51.60 | 34 | PHAGE_Klebsi_4LV2017_NC_047818 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega-Balleza, J.L.; Guerrero, A.; Castro-Escarpulli, G.; Martínez-Vázquez, A.V.; Cruz-Hernández, M.A.; Luna-Santillana, E.d.J.d.; Acosta-Cruz, E.; Rodríguez-Sánchez, I.P.; Rivera, G.; Bocanegra-García, V. Genomic Analysis of Multidrug-Resistant Escherichia coli Strains Isolated in Tamaulipas, Mexico. Trop. Med. Infect. Dis. 2023, 8, 458. https://doi.org/10.3390/tropicalmed8100458
Ortega-Balleza JL, Guerrero A, Castro-Escarpulli G, Martínez-Vázquez AV, Cruz-Hernández MA, Luna-Santillana EdJd, Acosta-Cruz E, Rodríguez-Sánchez IP, Rivera G, Bocanegra-García V. Genomic Analysis of Multidrug-Resistant Escherichia coli Strains Isolated in Tamaulipas, Mexico. Tropical Medicine and Infectious Disease. 2023; 8(10):458. https://doi.org/10.3390/tropicalmed8100458
Chicago/Turabian StyleOrtega-Balleza, Jessica L., Abraham Guerrero, Graciela Castro-Escarpulli, Ana Verónica Martínez-Vázquez, María Antonia Cruz-Hernández, Erick de Jesús de Luna-Santillana, Erika Acosta-Cruz, Irám Pablo Rodríguez-Sánchez, Gildardo Rivera, and Virgilio Bocanegra-García. 2023. "Genomic Analysis of Multidrug-Resistant Escherichia coli Strains Isolated in Tamaulipas, Mexico" Tropical Medicine and Infectious Disease 8, no. 10: 458. https://doi.org/10.3390/tropicalmed8100458
APA StyleOrtega-Balleza, J. L., Guerrero, A., Castro-Escarpulli, G., Martínez-Vázquez, A. V., Cruz-Hernández, M. A., Luna-Santillana, E. d. J. d., Acosta-Cruz, E., Rodríguez-Sánchez, I. P., Rivera, G., & Bocanegra-García, V. (2023). Genomic Analysis of Multidrug-Resistant Escherichia coli Strains Isolated in Tamaulipas, Mexico. Tropical Medicine and Infectious Disease, 8(10), 458. https://doi.org/10.3390/tropicalmed8100458












