Epidemiology of Community-Acquired Respiratory Tract Infections in Patients Admitted at the Emergency Departments
Abstract
:1. Introduction
2. Methods
Molecular Experiments
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Verani, J.R.; McCracken, J.; Arvelo, W.; Estevez, A.; Lopez, M.R.; Reyes, L.; Moir, J.C.; Bernart, C.; Moscoso, F.; Gray, J.; et al. Surveillance for hospitalized acute respiratory infection in Guatemala. PLoS ONE 2013, 8, e83600. [Google Scholar] [CrossRef]
- Stein, R.T.; Marostica, P.J. Community-acquired pneumonia: A review and recent advances. Pediatr. Pulmonol. 2007, 42, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Abeele, A.M.; Cartuyvels, R. Timely diagnosis of respiratory tract infections: Evaluation of the performance of the Respifinder assay compared to the xTAG respiratory viral panel assay. J. Clin. Virol. 2011, 52, 314–316. [Google Scholar] [CrossRef]
- Gonzales, R.; Malone, D.C.; Maselli, J.H.; Sande, M.A. Excessive antibiotic use for acute respiratory infections in the United States. Clin. Infect. Dis. 2001, 33, 757–762. [Google Scholar] [CrossRef]
- Fox, J.D. Respiratory virus surveillance and outbreak investigation. J. Clin. Virol. 2007, 40 (Suppl. S1), S24–S30. [Google Scholar] [CrossRef]
- Loeffelholz, M.J.; Pong, D.L.; Pyles, R.B.; Xiong, Y.; Miller, A.L.; Bufton, K.K.; Chonmaitree, T. Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J. Clin. Microbiol. 2011, 49, 4083–4088. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.H.; Zick, B.L.; Tsalik, E.L. Host-Based Diagnostics for acute respiratory infections. Clin. Ther. 2019, 41, 1923–1938. [Google Scholar] [CrossRef] [PubMed]
- Burnham, J.P.; Olsen, M.A.; Kollef, M.H. Re-estimating annual deaths due to multidrug-resistant organism infections. Infect Control. Hosp. Epidemiol. 2019, 40, 112–113. [Google Scholar] [CrossRef]
- Fleming-Dutra, K.E.; Hersh, A.L.; Shapiro, D.J.; Bartoces, M.; Enns, E.A.; File, T.M., Jr.; Finkelstein, J.A.; Gerber, J.S.; Hyun, D.Y.; Linder, J.A.; et al. Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010–2011. JAMA 2016, 315, 1864–1873. [Google Scholar] [CrossRef]
- Donnelly, J.P.; Baddley, J.W.; Wang, H.E. Antibiotic utilization for acute respiratory tract infections in US emergency departments. Antimicrob. Agents Chemother. 2014, 58, 1451–1457. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Park, J.Y.; Song, Y.; How, S.H.; Jung, K.S. Respiratory Infections Assembly of the APSR. Emerging respiratory infections threatening public health in the Asia-Pacific region: A position paper of the Asian Pacific Society of Respirology. Respirology 2019, 24, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Liapikou, A.; Cilloniz, C.; Mensa, J.; Torres, A. New antimicrobial approaches to gram positive respiratory infections. Pulm. Pharm. 2015, 32, 137–143. [Google Scholar] [CrossRef]
- Bulla, A.; Hitze, K.L. Acute respiratory infections: A review. Bull. World Health Organ. 1978, 56, 481–498. [Google Scholar] [PubMed]
- Pérez-Ruiz, M.; Pedrosa-Corral, I.; Sanbonmatsu-Gámez, S.; Navarro-Marí, M. Laboratory detection of respiratory viruses by automated techniques. Open Virol. J. 2012, 6, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Renaud, C.; Crowley, J.; Jerome, K.R.; Kuypers, J. Comparison of FilmArray Respiratory Panel and laboratory-developed real-time reverse transcription-polymerase chain reaction assays for respiratory virus detection. Diagn. Microbiol. Infect Dis. 2012, 74, 379–383. [Google Scholar] [CrossRef]
- Fox, J.D. Nucleic acid amplification tests for detection of respiratory viruses. J. Clin. Virol. 2007, 40 (Suppl. S1), S15–S23. [Google Scholar] [CrossRef]
- Bashir, U.; Alam, M.M.; Sadia, H.; Zaidi, S.S.Z.; Kazi, B.M. Molecular characterization of circulating respiratory syncytial virus (RSV) genotypes in Gilgit Baltistan Province of Pakistan during 2011-2012 winter season. PLoS ONE 2013, 8, e74018. [Google Scholar] [CrossRef]
- Bashir, U.; Nisar, N.; Arshad, Y.; Alam, M.M.; Ashraf, A.; Sadia, H.; Kazi, B.M.; Zaidi, S.S.Z. Respiratory syncytial virus and influenza are the key viral pathogens in children <2 years hospitalized with bronchiolitis and pneumonia in Islamabad Pakistan. Arch. Virol. 2017, 162, 763–773. [Google Scholar] [CrossRef]
- Badar, N.; Bashir Aamir, U.; Mehmood, M.R.; Nisar, N.; Alam, M.M.; Kazi, B.M.; Zaidi, S.S.Z. Influenza virus surveillance in Pakistan during 2008-2011. PLoS ONE 2013, 8, e79959. [Google Scholar] [CrossRef]
- Tsang, K.W.; File, T.M., Jr. Respiratory infections unique to Asia. Respirology 2008, 13, 937–949. [Google Scholar] [CrossRef]
- Matsushima, T.; Miyashita, N.; File, T.M., Jr. Etiology and management of community-acquired pneumonia in Asia. Curr. Opin. Infect Dis. 2002, 15, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Oh, W.S.; Kang, C.I.; Chung, D.R.; Peck, K.R.; Ko, K.S.; Yeom, J.S.; Kim, C.K.; Kim, S.W.; Chang, H.-H.; et al. Epidemiology and clinical outcomes of community-acquired pneumonia in adult patients in Asian countries: A prospective study by the Asian network for surveillance of resistant pathogens. Int. J. Antimicrob. Agents 2008, 31, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Robert, L.; Mark, V.; Moayed, A.; Nivashen, A.; Vinod, R.; Sophie, P.; Mohamed, E.W.; Rusheng, C. Antimicrobial prescribing and outcomes of community-acquired pneumonia in Australian hospitalized patients: A cross-sectional study. J. Int. Med. Res. 2021, 49, 3000605211058366. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.C.; Huah, L.W.; Lin, J.T.; Goh, A.; Ling, H.; Moh, C.O. Lower respiratory tract infection in hospitalized children. Respirology 2003, 8, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Mandell, L.A.; Wunderink, R.G.; Anzueto, A.; Bartlett, J.G.; Campbell, G.D.; Dean, N.C.; Dowell, S.F.; File, T.M., Jr.; Musher, D.M.; Niederman, M.S.; et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 2007, 44 (Suppl. S2), S27–S72. [Google Scholar] [CrossRef]
- Cillóniz, C.; Cardozo, C.; García-Vidal, C. Epidemiology, pathophysiology, and microbiology of community-acquired pneumonia. Ann. Res. Hosp. 2018, 2, 1. [Google Scholar] [CrossRef]
- Alimi, Y.; Lim, W.S.; Lansbury, L.; Leonardi-Bee, J.; Nguyen-Van-Tam, J.S. Systematic review of respiratory viral pathogens identified in adults with community-acquired pneumonia in Europe. J. Clin. Virol. 2017, 95, 26–35. [Google Scholar] [CrossRef]
- Lieberman, D.; Lieberman, D.; Korsonsky, I.; Ben-Yaakov, M.; Lazarovich, Z.; Friedman, M.G.; Dvoskin, B.; Leinonen, M.; Ohana, B.; Boldur, I. A comparative study of the etiology of adult upper and lower respiratory tract infections in the community. Diagn. Microbiol. Infect Dis. 2002, 42, 21–28. [Google Scholar] [CrossRef]
- Shi, T.; Arnott, A.; Semogas, I.; Falsey, A.R.; Openshaw, P.; Wedzicha, J.; Campbell, H.; Nair, H.; Zhang, S.; Li, Y.; et al. The etiological role of common respiratory viruses in acute respiratory infections in older adults: A systematic review and meta-analysis. J. Infect Dis. 2020, 222 (Suppl. S7), S563–S569. [Google Scholar] [CrossRef]
- Tsolia, M.N.; Psarras, S.; Bossios, A.; Audi, H.; Paldanius, M.; Gourgiotis, D.; Kallergi, K.; Kafetzis, D.A.; Constantopoulos, A.; Papadopoulos, N. Etiology of community-acquired pneumonia in hospitalized school-age children: Evidence for high prevalence of viral infections. Clin. Infect Dis. 2004, 39, 681–686. [Google Scholar] [CrossRef] [Green Version]
- Masse, S.; Capai, L.; Falchi, A. Epidemiology of respiratory pathogens among elderly nursing home residents with acute respiratory infections in Corsica, France, 2013–2017. Biomed Res. Int. 2017, 2017, 1423718. [Google Scholar] [CrossRef] [PubMed]
- Garibaldi, R.A. Epidemiology of community-acquired respiratory tract infections in adults. Incidence, etiology, and impact. Am. J. Med. 1985, 78, 32–37. [Google Scholar] [CrossRef]
- Al-Ali, M.K.; Batchoun, R.G.; Al-Nour, T.M. Etiology of community-acquired pneumonia in hospitalized patients in Jordan. Saudi. Med. J. 2006, 27, 813–816. [Google Scholar] [PubMed]
- Aston, S.J.; Ho, A.; Jary, H.; Huwa, J.; Mitchell, T.; Ibitoye, S.; Greenwood, S.; Joekes, E.; Daire, A.; Mallewa, J.; et al. Etiology and risk factors for mortality in an adult community-acquired pneumonia cohort in Malawi. Am. J. Respir. Crit. Care Med. 2019, 200, 359–369. [Google Scholar] [CrossRef]
- Ozyilmaz, E.; Akan, O.A.; Gulhan, M.; Ahmed, K.; Nagatake, T. Major bacteria of community-acquired respiratory tract infections in Turkey. Jpn. J. Infect Dis. 2005, 58, 50–52. [Google Scholar]
- Marrie, T.J.; Poulin-Costello, M.; Beecroft, M.D.; Herman-Gnjidic, Z. Etiology of community-acquired pneumonia treated in an ambulatory setting. Respir. Med. 2005, 99, 60–65. [Google Scholar] [CrossRef]
- Woodhead, M. Community-acquired pneumonia in Europe: Causative pathogens and resistance patterns. Eur. Respir. J. Suppl. 2002, 36, 20s–27s. [Google Scholar] [CrossRef]
- Vaccine Uptake in Canadian Adults: Highlights from the 2016 Adult National Immunization Coverage Survey (aNICS)-Canada.ca. Available online: https://www.canada.ca/en/services/health/publications/healthy-living/2016-vaccine-uptake-canadian-adults-survey.html (accessed on 12 April 2021).
- Lupisan, S.; Suzuki, A.; Macalalad, N.; Egos, R.; Sombrero, L.; Okamoto, M.; Dapat, C.; Mondoy, M.; Galang, H.; Zeta, V.F.F.; et al. Etiology and epidemiology of community-acquired pneumonia in adults requiring hospital admission: A prospective study in rural Central Philippines. Int. J. Infect Dis. 2019, 80, 46–53. [Google Scholar] [CrossRef]
- Ziko, L.M.; Hoffman, T.W.; Fwoloshi, S.; Chanda, D.; Nampungwe, Y.M.; Patel, D.; Bobat, H.; Moonga, A.; Chirwa, L.; Hachaambwa, L.; et al. Aetiology and prognosis of community-acquired pneumonia at the Adult University Teaching Hospital in Zambia. PLoS ONE 2022, 17, e0271449. [Google Scholar] [CrossRef]
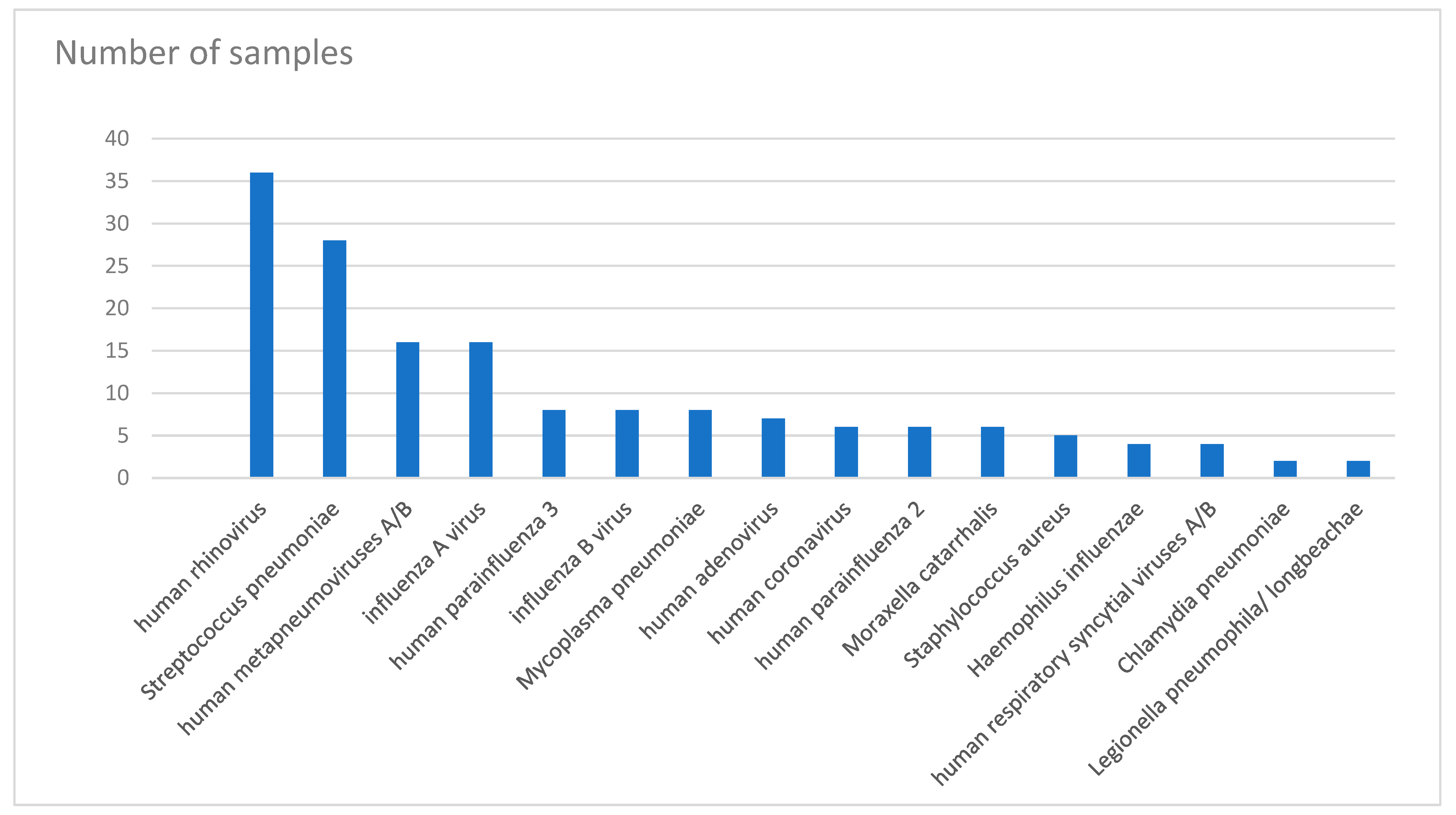

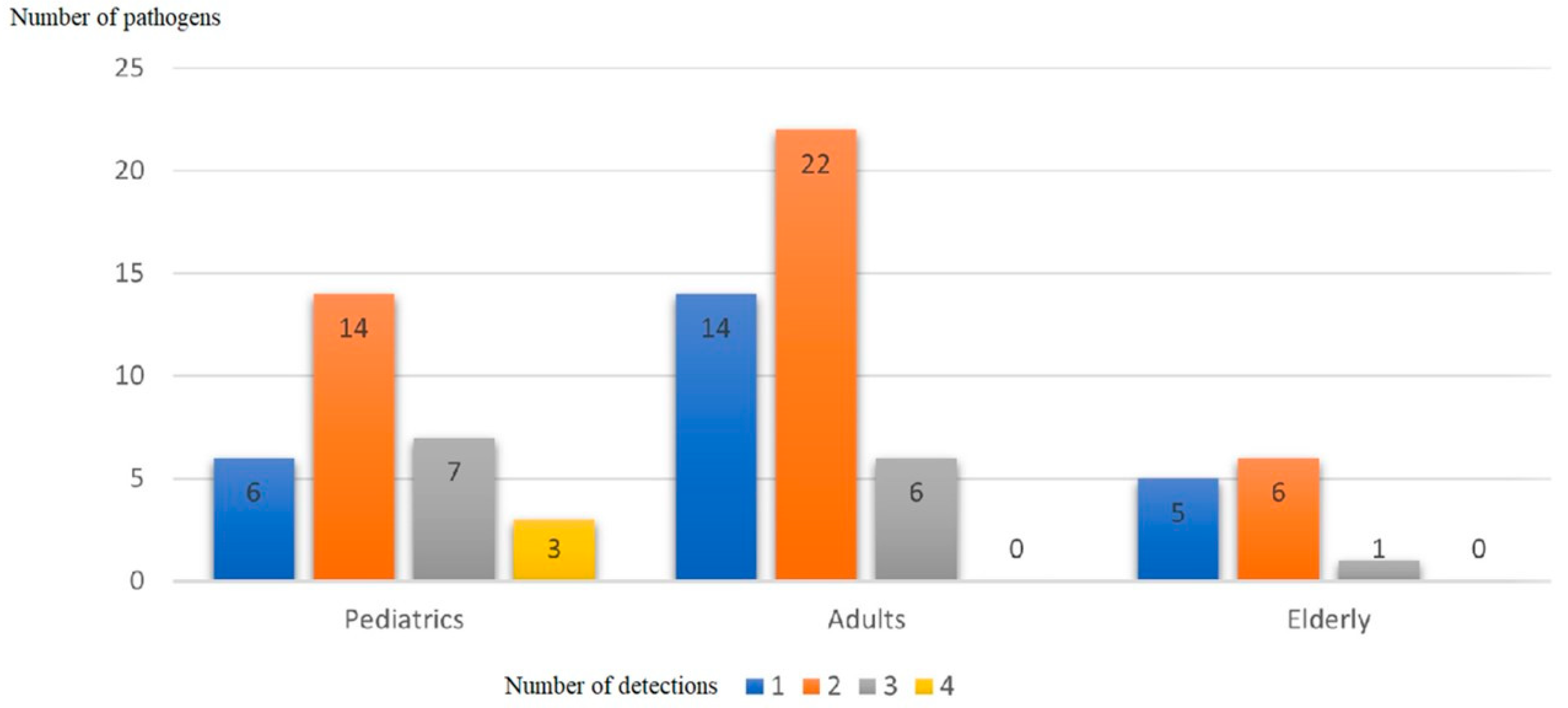
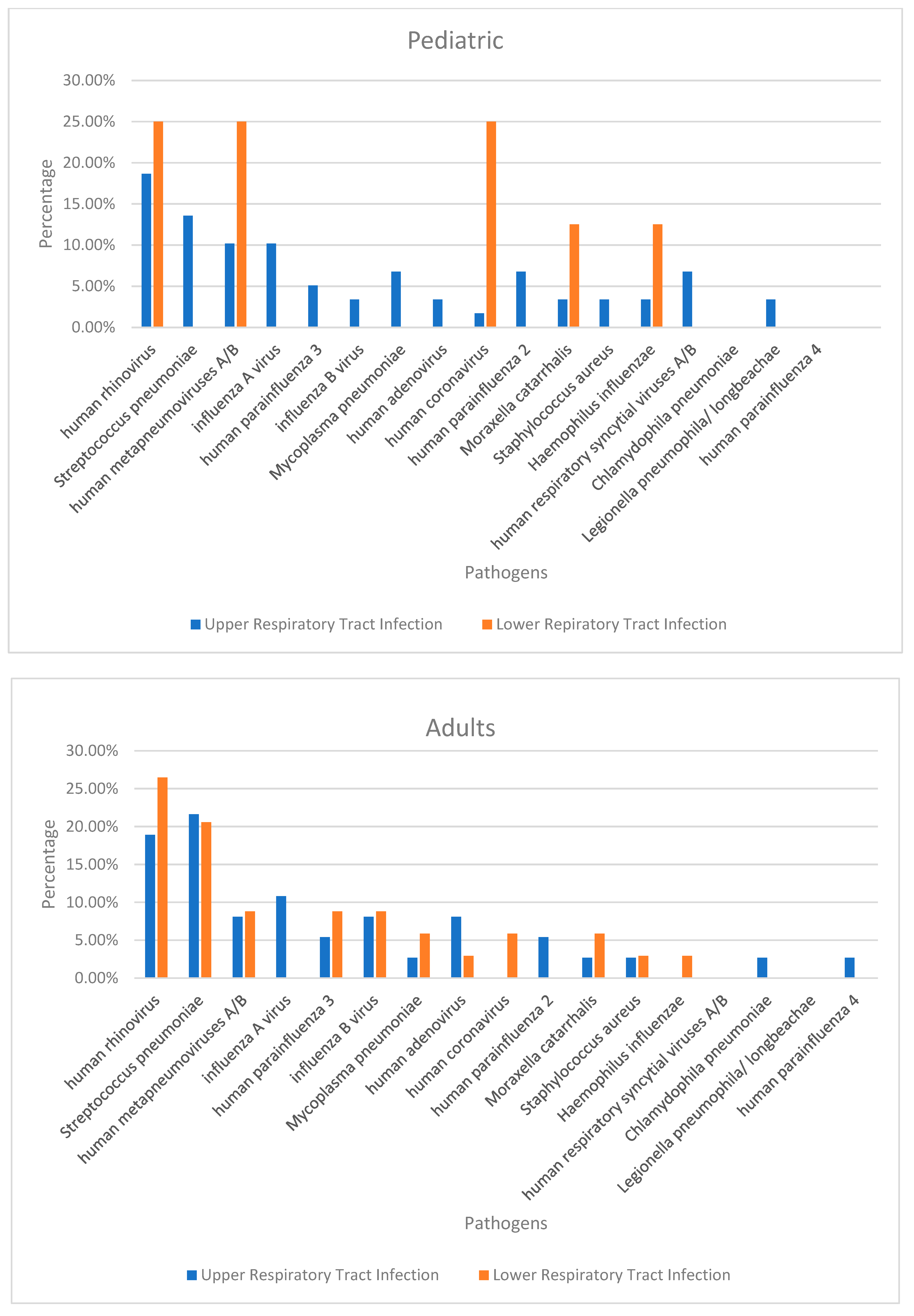
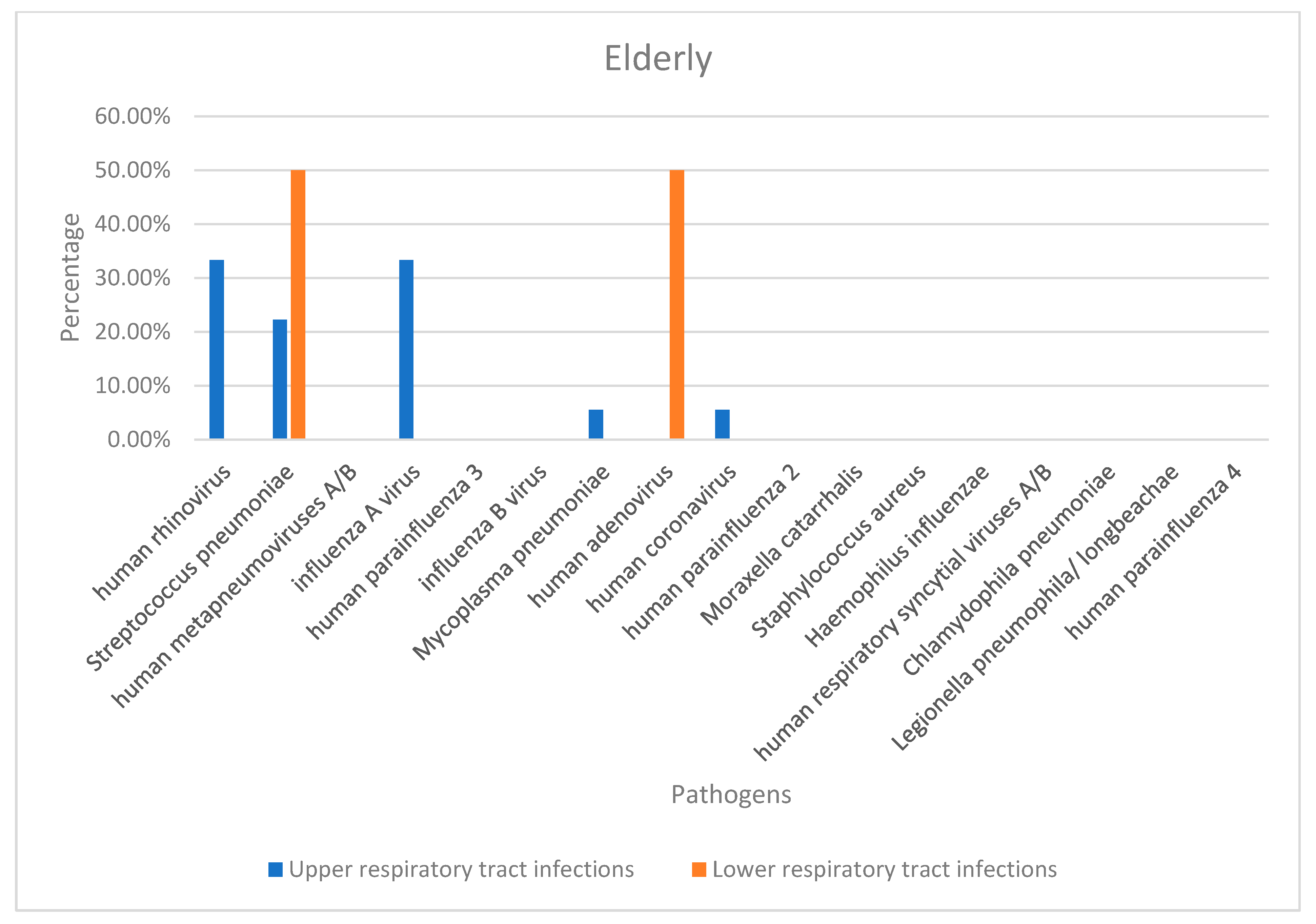
| Pathogen | Total Number | Number of Detections | |||
|---|---|---|---|---|---|
| 1 detection | 2 detections | 3 detections | 4 detections | ||
| human rhinovirus | 36 | 2 | 23 | 8 | 3 |
| Streptococcus pneumoniae | 28 | 6 | 16 | 4 | 2 |
| human metapneumoviruses A/B | 16 | 5 | 5 | 5 | 1 |
| influenza A virus | 16 | 6 | 7 | 3 | 0 |
| human parainfluenza 3 | 8 | 1 | 2 | 4 | 1 |
| influenza B virus | 8 | 3 | 4 | 1 | 0 |
| Mycoplasma pneumoniae | 8 | 2 | 3 | 3 | 0 |
| human adenovirus | 7 | 0 | 4 | 3 | 0 |
| human coronavirus | 6 | 0 | 3 | 2 | 1 |
| human parainfluenza 2 | 6 | 0 | 1 | 4 | 1 |
| Moraxella catarrhalis | 6 | 0 | 3 | 2 | 1 |
| Staphylococcus aureus | 5 | 0 | 3 | 1 | 1 |
| Haemophilus influenzae | 4 | 0 | 2 | 2 | 0 |
| human respiratory syncytial viruses A/B | 4 | 0 | 2 | 1 | 1 |
| Chlamydia pneumoniae | 2 | 0 | 1 | 1 | 0 |
| Legionella pneumophila/longbeachae | 2 | 0 | 2 | 0 | 0 |
| human parainfluenza 4 | 1 | 0 | 1 | 0 | 0 |
| Aetiology of Respiratory Tract Infections | ||||||||
|---|---|---|---|---|---|---|---|---|
| Source | Country | Sampling Technique | Upper Respiratory Tract Infections | Lower Respiratory Tract Infections | ||||
| Elderly | ADULTS | Pediatric | Adults | |||||
| Viral Aetiology | Bacterial Aetiology | Viral Artiology | Bacterial Aetiology | Bacterial Aetiology | Viral Aetiology | |||
| [32] | USA | not mentioned | Presumed virus or chlamydia 30–40% (not tested) | Streptooccus pneumonia | ||||
| Group A Streptococci 5–10% | rhinovirus 25–30% | Enteric gram-negative organisms | ||||||
| Mycoplasma 5–10% | coronavirus > 10% | Staphylococcus aureus | ||||||
| Influenza Virus, RSV, Adenovirus and Parainfluenza Virus 10–15% | Hemophilus influenza | |||||||
| Other viruses | Pseudomonas aeruginosa | |||||||
| [36] | Canada | Blood culture | Unknown 51.6% | |||||
| sputum culture | Mycoplasma pneumoniae 15% | |||||||
| acute and convalescent serum samples for serology | Chlamydia pneumoniae 12% | |||||||
| Antibodies to Mycoplasma pneumoniae and Chlamydia pneumoniae determined using enzyme-linked immunosorbent assays | Streptococcus pneumoniae 5.9% | |||||||
| Haemophilus influenzae 4.9% | ||||||||
| Chlamydia pneumoniae and Mycoplasma pneumoniae 2.1% | ||||||||
| Haemophilus parainfluenzae 1.9% | ||||||||
| Staphylococcus aureus 1.1% | ||||||||
| Moraxella catarrhalis 1.1% | ||||||||
| Streptococcus species 0.9% | ||||||||
| Other 2.8% | ||||||||
| [37] | Europe (UK, Spain and Sweden) | not mentioned | No pathogen identified 49.8% | viruses 11.7% | ||||
| Streptococcus pneumoniae 19.3% | ||||||||
| Mycoplasma pneumoniae 11.1% | ||||||||
| Chlamydia pneumoniae 8% | ||||||||
| Haemophilus influenzae 3.3% | ||||||||
| [31] | France | QiaAmp MinElute virus spin kits | Influenza A (H3N2) | |||||
| real-time Reverse Transcription quantitative PCR (RT-qPCR) | Human rhinovirus 16% | |||||||
| Human coronavirus OC43 7% | ||||||||
| Respiratory Syncytial Virus 5% | ||||||||
| Human metapneumovirus 5% | ||||||||
| Influenza B/Victoria 5% | ||||||||
| [34] | Malawi | blood culture | No pathogen detected 39.4% | Influenza viruses 8.8% | ||||
| Streptococcus pneumoniae urinary antigen detection | Mycobacterium tuberculosis 23% | Adenovirus 7.7% | ||||||
| sputum mycobacterial culture | Streptococcus pneumoniae 21.4% | Coronaviruses 6.8% | ||||||
| Xpert MTB/RIF | Nontuberculous mycobacteria 2.9% | Parainfluenza viruses 3.7% | ||||||
| nasopharyngeal aspirate multiplex PCR | Salmonella enterica serovar Typhi 2.2% | Rhinovirus 4.2% | ||||||
| Nontyphoidal Salmonella 1.6% | Bocavirus 2.9% | |||||||
| Mycoplasma pneumoniae 1.3% | Metapneumovirus 2.0% | |||||||
| Other gram-negative enteric bacilli 0.7% | RSV 1.8% | |||||||
| Staphylococcus aureus 0.4% | Enterovirus 1.1% | |||||||
| Chlamydia pneumoniae 0.4% | Parechovirus 1.1% | |||||||
| [35] | Turkey | sputum cultures | Haemophilus influenzae 44.9% | |||||
| Streptococcus pneumoniae 25.5% | ||||||||
| Moraxella catarrhalis 12.2% | ||||||||
| Pseudomonas aeruginosa 3.1% | ||||||||
| Klebsiella spp. 1% | ||||||||
| Haemophilus parainfluenzae 1% | ||||||||
| Staphylococcus aureus 1% | ||||||||
| [33] | Jordan | sputum cultures | Streptococcus pneumoniae 26% | Chlamydia pneumoniae 14% | ||||
| Chlamydia pneumoniae 23% | Mycoplasma pneumoniae 6% | |||||||
| Haemophilus influenzae 17% | Streptococcus pneumoniae 3% | |||||||
| Mycoplasma pneumoniae 9% | Haemophilus influenzae 3% | |||||||
| Legionella pneumophila 6% | Pseudomonas aeruginosa 3% | |||||||
| Klebsiella pneumoniae 6% | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helou, M.; Mahdi, A.; Daoud, Z.; Mokhbat, J.; Farra, A.; Nassar, E.; Nehme, R.; Abboud, E.; Masri, K.; Husni, R. Epidemiology of Community-Acquired Respiratory Tract Infections in Patients Admitted at the Emergency Departments. Trop. Med. Infect. Dis. 2022, 7, 233. https://doi.org/10.3390/tropicalmed7090233
Helou M, Mahdi A, Daoud Z, Mokhbat J, Farra A, Nassar E, Nehme R, Abboud E, Masri K, Husni R. Epidemiology of Community-Acquired Respiratory Tract Infections in Patients Admitted at the Emergency Departments. Tropical Medicine and Infectious Disease. 2022; 7(9):233. https://doi.org/10.3390/tropicalmed7090233
Chicago/Turabian StyleHelou, Mariana, Ahmad Mahdi, Ziad Daoud, Jacques Mokhbat, Anna Farra, Elma Nassar, Ralph Nehme, Edmond Abboud, Khalil Masri, and Rola Husni. 2022. "Epidemiology of Community-Acquired Respiratory Tract Infections in Patients Admitted at the Emergency Departments" Tropical Medicine and Infectious Disease 7, no. 9: 233. https://doi.org/10.3390/tropicalmed7090233
APA StyleHelou, M., Mahdi, A., Daoud, Z., Mokhbat, J., Farra, A., Nassar, E., Nehme, R., Abboud, E., Masri, K., & Husni, R. (2022). Epidemiology of Community-Acquired Respiratory Tract Infections in Patients Admitted at the Emergency Departments. Tropical Medicine and Infectious Disease, 7(9), 233. https://doi.org/10.3390/tropicalmed7090233





