Different In Vitro Drug Susceptibility Profile of Plasmodium falciparum Isolates from Two Adjacent Areas of Northeast Myanmar and Molecular Markers for Drug Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Parasite Sample Collection
2.2. Parasite Culture and Drug Assays
2.3. Ring Survival Assay (RSA)
2.4. Sequencing of Genes Associated with Drug Resistance
2.5. Statistical Analyses
3. Results
3.1. In Vitro Drug Susceptibilities of P. falciparum from the China–Myanmar Border
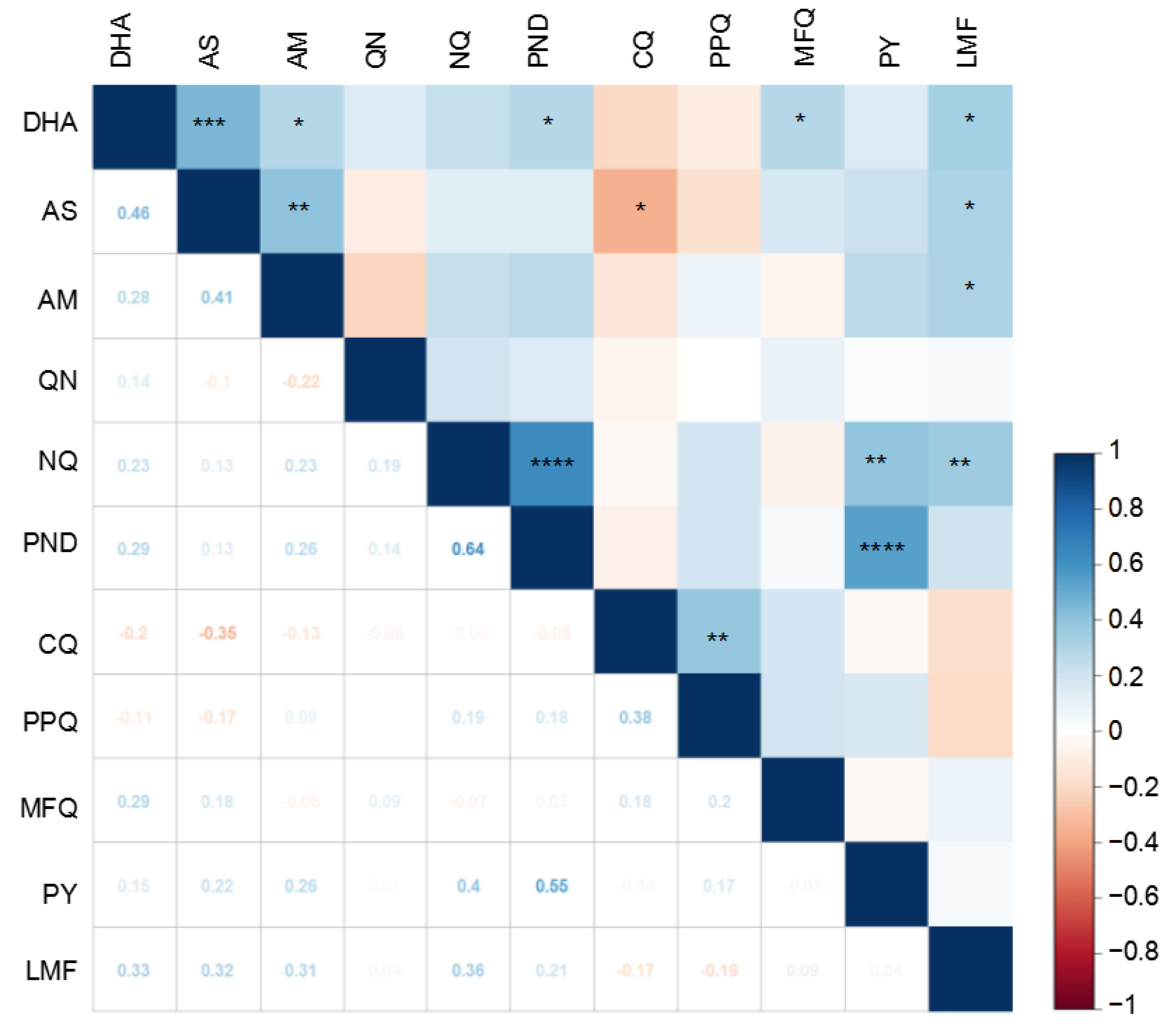
3.2. Correlations between Susceptibilities to Different Drugs
3.3. Polymorphisms in Genes Associated with Drug Resistance
3.4. Multidrug Resistance Haplotypes
3.5. Regional Differences in Drug Susceptibility and Genetic Polymorphisms
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaehler, N.; Adhikari, B.; Cheah, P.Y.; von Seidlein, L.; Day, N.P.J.; Paris, D.H.; Tanner, M.; Pell, C. Prospects and strategies for malaria elimination in the Greater Mekong Sub-region: A qualitative study. Malar. J. 2019, 18, 203. [Google Scholar] [CrossRef] [PubMed]
- WHO|Regional Office for the Western Pacific. Strategy for Malaria Elimination in the Greater Mekong Subregion: 2015–2030. Available online: https://www.who.int/publications/i/item/9789290617181 (accessed on 21 May 2015).
- Zhou, X.N. China declared malaria-free: A milestone in the world malaria eradication and Chinese public health. Infect. Dis. Poverty 2021, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Yan, G.; Sattabongkot, J.; Cao, Y.; Chen, B.; Chen, X.; Fan, Q.; Fang, Q.; Jongwutiwes, S.; Parker, D.; et al. Malaria in the Greater Mekong Subregion: Heterogeneity and complexity. Acta Trop. 2012, 121, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Malla, P.; Zhang, J.; Xu, S.; Li, C.; Zhao, Y.; Wang, Q.; Kyaw, M.P.; Cao, Y.; Yang, Z.; et al. Increasing trends of malaria in a border area of the Greater Mekong Subregion. Malar. J. 2019, 18, 309. [Google Scholar] [CrossRef] [PubMed]
- Lo, E.; Lam, N.; Hemming-Schroeder, E.; Nguyen, J.; Zhou, G.; Lee, M.C.; Yang, Z.; Cui, L.; Yan, G. Frequent Spread of Plasmodium vivax Malaria Maintains High Genetic Diversity at the Myanmar-China Border, without Distance and Landscape Barriers. J. Infect. Dis. 2017, 216, 1254–1263. [Google Scholar] [CrossRef]
- He, X.; Zhong, D.; Zou, C.; Pi, L.; Zhao, L.; Qin, Y.; Pan, M.; Wang, S.; Zeng, W.; Xiang, Z.; et al. Unraveling the Complexity of Imported Malaria Infections by Amplicon Deep Sequencing. Front Cell. Infect. Microbiol. 2021, 11, 725859. [Google Scholar] [CrossRef]
- Fairhurst, R.M.; Dondorp, A.M. Artemisinin-Resistant Plasmodium falciparum malaria. Microbiol. Spectr. 2016, 4, 10. [Google Scholar] [CrossRef]
- Zeng, W.; Bai, Y.; Wang, M.; Wang, Z.; Deng, S.; Ruan, Y.; Feng, S.; Yang, Z.; Cui, L. Significant Divergence in Sensitivity to Antimalarial Drugs between Neighboring Plasmodium falciparum Populations along the Eastern Border of Myanmar. Antimicrob. Agents Chemother. 2017, 61, e01689-16. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Z.; Yuan, L.; Zhou, G.; Parker, D.; Lee, M.C.; Yan, G.; Fan, Q.; Xiao, Y.; Cao, Y.; et al. Clinical Efficacy of Dihydroartemisinin-Piperaquine for the Treatment of Uncomplicated Plasmodium falciparum Malaria at the China-Myanmar Border. Am. J. Trop. Med. Hyg. 2015, 93, 577–583. [Google Scholar] [CrossRef]
- Malaria Clinical Management Guidelines, Ministry of Health, Myanmar. Available online: https://moh.nugmyanmar.org/wp-content/uploads/2022/07/Malaria_Clinical_Management_Guidelines__June_2022___Ver_1__MOH_NUG_of_Myanmar.pdf (accessed on 28 November 2022).
- Myint, M.K.; Rasmussen, C.; Thi, A.; Bustos, D.; Ringwald, P.; Lin, K. Therapeutic efficacy and artemisinin resistance in northern Myanmar: Evidence from in vivo and molecular marker studies. Malar. J. 2017, 16, 143. [Google Scholar] [CrossRef]
- Meng, H.; Zhang, R.; Yang, H.; Fan, Q.; Su, X.; Miao, J.; Cui, L.; Yang, Z. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China-Myanmar border area to quinine and association with polymorphism in the NA+/H+ exchanger. Antimicrob. Agents Chemother. 2010, 54, 4306–4313. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Trager, W.; Jensen, J.B. Human malaria parasites in continuous culture. Science. 1976, 193, 673–675. [Google Scholar] [CrossRef] [PubMed]
- Smilkstein, M.; Sriwilaijaroen, N.; Kelly, J.X.; Wilairat, P.; Riscoe, M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 2004, 48, 1803–1806. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, F.A.; Cabrera, M.; Wang, M.; Brashear, A.; Kemirembe, K.; Wang, Z.; Miao, J.; Chookajorn, T.; Yang, Z.; Cao, Y.; et al. Plasmodium falciparum falcipain-2a polymorphisms in Southeast Asia and Their Association with Artemisinin Resistance. J. Infect. Dis. 2018, 218, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, B.; Amaratunga, C.; Khim, N.; Sreng, S.; Chim, P.; Kim, S.; Lim, P.; Mao, S.; Sopha, C.; Sam, B.; et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: In-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 2013, 13, 1043–1049. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, G.H.; Zou, C.Y.; Su, P.C.; Liu, H.E.; Yang, Z.Q. Overview of the improvement of the ring-stage survival assay-a novel phenotypic assay for the detection of artemisinin-resistant Plasmodium falciparum. Zool. Res. 2017, 38, 317–320. [Google Scholar]
- Amaratunga, C.; Witkowski, B.; Dek, D.; Try, V.; Khim, N.; Miotto, O.; Menard, D.; Fairhurst, R.M. Plasmodium falciparum founder populations in western Cambodia have reduced artemisinin sensitivity in vitro. Antimicrob. Agents Chemother. 2014, 58, 4935–4937. [Google Scholar] [CrossRef]
- Foguim, F.T.; Bogreau, H.; Gendrot, M.; Mosnier, J.; Fonta, I.; Benoit, N.; Amalvict, R.; Madamet, M.; Wein, S.; Pradines, B.; et al. Prevalence of mutations in the Plasmodium falciparum chloroquine resistance transporter, Pfcrt, and association with ex vivo susceptibility to common anti-malarial drugs against African Plasmodium falciparum isolates. Malar. J. 2020, 19, 201. [Google Scholar] [CrossRef]
- Yang, Z.; Li, C.; Miao, M.; Zhang, Z.; Sun, X.; Meng, H.; Li, J.; Fan, Q.; Cui, L. Multidrug-resistant genotypes of Plasmodium falciparum, Myanmar. Emerg. Infect. Dis. 2011, 17, 498–501. [Google Scholar] [CrossRef]
- Wang, Z.; Shrestha, S.; Li, X.; Miao, J.; Yuan, L.; Cabrera, M.; Grube, C.; Yang, Z.; Cui, L. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007–2012. Malar. J. 2015, 14, 168. [Google Scholar] [CrossRef]
- Gupta, B.; Xu, S.; Wang, Z.; Sun, L.; Miao, J.; Cui, L.; Yang, Z. Plasmodium falciparum multidrug resistance protein 1 (pfmrp1) gene and its association with in vitro drug susceptibility of parasite isolates from north-east Myanmar. J. Antimicrob. Chemother. 2014, 69, 2110–2117. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ringwald, P.; Bickii, J.; Basco, L.K. In vitro activity of antimalarials against clinical isolates of Plasmodium falciparum in Yaounde, Cameroon. Am. J. Trop. Med. Hyg. 1996, 55, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Pradines, B.; Tall, A.; Parzy, D.; Spiegel, A.; Fusai, T.; Hienne, R.; Trape, J.F.; Doury, J.C. In-vitro activity of pyronaridine and amodiaquine against African isolates (senegal) of Plasmodium falciparum in comparison with standard antimalarial agents. J. Antimicrob. Chemother. 1998, 42, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Pradines, B.; Tall, A.; Fusai, T.; Spiegel, A.; Hienne, R.; Rogier, C.; Trape, J.F.; Le Bras, J.; Parzy, D. In vitro activities of benflumetol against 158 Senegalese isolates of Plasmodium falciparum in comparison with those of standard antimalarial drugs. Antimicrob. Agents Chemother. 1999, 43, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Aubouy, A.; Jafari, S.; Huart, V.; Migot-Nabias, F.; Mayombo, J.; Durand, R.; Bakary, M.; Le Bras, J.; Deloron, P. DHFR and DHPS genotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine-pyrimethamine treatment efficacy. J. Antimicrob. Chemother. 2003, 52, 43–49. [Google Scholar] [CrossRef]
- Hao, M.; Jia, D.; Li, Q.; He, Y.; Yuan, L.; Xu, S.; Chen, K.; Wu, J.; Shen, L.; Sun, L.; et al. In vitro sensitivities of Plasmodium falciparum isolates from the China-Myanmar border to piperaquine and association with polymorphisms in candidate genes. Antimicrob. Agents Chemother. 2013, 57, 1723–1729. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, J.; Geng, J.; Xu, S.; Deng, S.; Zeng, W.; Wang, Z.; NgassaMbenda, H.G.; Zhang, J.; Li, N.; et al. Longitudinal surveillance of drug resistance in Plasmodium falciparum isolates from the China-Myanmar border reveals persistent circulation of multidrug resistant parasites. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 320–328. [Google Scholar] [CrossRef]
- Cheruiyot, J.; Ingasia, L.A.; Omondi, A.A.; Juma, D.W.; Opot, B.H.; Ndegwa, J.M.; Mativo, J.; Cheruiyot, A.C.; Yeda, R.; Okudo, C.; et al. Polymorphisms in Pfmdr1, Pfcrt, and Pfnhe1 genes are associated with reduced in vitro activities of quinine in plasmodium falciparum isolates from western Kenya. Antimicrob. Agents Chemother. 2014, 58, 3737–3743. [Google Scholar] [CrossRef]
- Ariey, F.; Witkowski, B.; Amaratunga, C.; Beghain, J.; Langlois, A.C.; Khim, N.; Kim, S.; Duru, V.; Bouchier, C.; Ma, L.; et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014, 505, 50–55. [Google Scholar] [CrossRef]
- Fidock, D.A.; Nomura, T.; Talley, A.K.; Cooper, R.A.; Dzekunov, S.M.; Ferdig, M.T.; Ursos, L.M.; Sidhu, A.B.; Naude, B.; Deitsch, K.W.; et al. Mutations in the P. falciparum digestive vacuole transmembrane protein Pfcrt and evidence for their role in chloroquine resistance. Mol. Cell 2000, 6, 861–871. [Google Scholar] [CrossRef]
- Happi, T.C.; Thomas, S.M.; Gbotosho, G.O.; Falade, C.O.; Akinboye, D.O.; Gerena, L.; Hudson, T.; Sowunmi, A.; Kyle, D.E.; Milhous, W.; et al. Point mutations in the pfcrt and pfmdr-1 genes of Plasmodium falciparum and clinical response to chloroquine, among malaria patients from Nigeria. Ann. Trop. Med. Parasitol. 2003, 97, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Khim, N.; Bouchier, C.; Ekala, M.T.; Incardona, S.; Lim, P.; Legrand, E.; Jambou, R.; Doung, S.; Puijalon, O.M.; Fandeur, T. Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob. Agents Chemother. 2005, 49, 3147–3152. [Google Scholar] [CrossRef] [PubMed]
- Iyer, J.K.; Milhous, W.K.; Cortese, J.F.; Kublin, J.G.; Plowe, C.V. Plasmodium falciparum cross-resistance between trimethoprim and pyrimethamine. Lancet 2001, 358, 1066–1067. [Google Scholar] [CrossRef] [PubMed]
- Sugaram, R.; Suwannasin, K.; Kunasol, C.; Mathema, V.B.; Day, N.P.J.; Sudathip, P.; Prempree, P.; Dondorp, A.M.; Imwong, M. Molecular characterization of Plasmodium falciparum antifolate resistance markers in Thailand between 2008 and 2016. Malar. J. 2020, 19, 107. [Google Scholar] [CrossRef]
- Menard, D.; Khim, N.; Beghain, J.; Adegnika, A.A.; Shafiul-Alam, M.; Amodu, O.; Rahim-Awab, G.; Barnadas, C.; Berry, A.; Boum, Y.; et al. A Worldwide Map of Plasmodium falciparum K13-Propeller Polymorphisms. N. Engl. J. Med. 2016, 374, 2453–2464. [Google Scholar] [CrossRef]
- Huang, F.; Takala-Harrison, S.; Jacob, C.G.; Liu, H.; Sun, X.; Yang, H.; Nyunt, M.M.; Adams, M.; Zhou, S.; Xia, Z.; et al. A Single Mutation in K13 Predominates in Southern China and Is Associated with Delayed Clearance of Plasmodium falciparum Following Artemisinin Treatment. J. Infect. Dis. 2015, 212, 1629–1635. [Google Scholar] [CrossRef]
- Tun, K.M.; Imwong, M.; Lwin, K.M.; Win, A.A.; Hlaing, T.M.; Hlaing, T.; Lin, K.; Kyaw, M.P.; Plewes, K.; Faiz, M.A.; et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: A cross-sectional survey of the K13 molecular marker. Lancet Infect. Dis. 2015, 15, 415–421. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.; Zhao, Y.; Ye, R.; Zhang, D.; Pan, W. Introduction of F446I mutation in the K13 propeller gene leads to increased ring survival rates in Plasmodium falciparum isolates. Malar. J. 2018, 17, 248. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Cabrera, M.; Zhang, Y.; Gupta, B.; Wu, Y.; Kemirembe, K.; Hu, Y.; Liang, X.; Brashear, A.; et al. Artemisinin resistance at the China-Myanmar border and association with mutations in the K13 propeller gene. Antimicrob. Agents Chemother. 2015, 59, 6952–6959. [Google Scholar] [CrossRef]
- Zhang, J.; Li, N.; Siddiqui, F.A.; Xu, S.; Geng, J.; Zhang, J.; He, X.; Zhao, L.; Pi, L.; Zhang, Y.; et al. In vitro susceptibility of Plasmodium falciparum isolates from the China-Myanmar border area to artemisinins and correlation with K13 mutations. Int. J. Parasitol. Drugs Drug Resist. 2019, 10, 20–27. [Google Scholar] [CrossRef]
- Siddiqui, F.A.; Boonhok, R.; Cabrera, M.; Mbenda, H.G.N.; Wang, M.; Min, H.; Liang, X.; Qin, J.; Zhu, X.; Miao, J.; et al. Role of Plasmodium falciparum Kelch 13 Protein Mutations in P. Falciparum Populations from Northeastern Myanmar in Mediating Artemisinin Resistance. mBio 2020, 11, e01134-19. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, L.C.; Singh, P.; Monteiro, W.M.; Magris, M.; Cox, H.; Lazrek, Y.; Melo, G.C.; Marchesini, P.; Alexandre, J.S.F.; Alvarez, A.M.; et al. Kelch13 mutations in Plasmodium falciparum and risk of spreading in Amazon basin countries. J. Antimicrob. Chemother. 2021, 76, 2854–2862. [Google Scholar] [CrossRef] [PubMed]
- Torrentino-Madamet, M.; Fall, B.; Benoit, N.; Camara, C.; Amalvict, R.; Fall, M.; Dionne, P.; Ba Fall, K.; Nakoulima, A.; Diatta, B.; et al. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar. J. 2014, 13, 472. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.D.; Bigira, V.; Kapisi, J.; Muhindo, M.; Kamya, M.R.; Havlir, D.V.; Dorsey, G.; Rosenthal, P.J. Polymorphisms in k13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS ONE 2014, 9, e105690. [Google Scholar] [CrossRef] [PubMed]
- Boussaroque, A.; Fall, B.; Madamet, M.; Camara, C.; Benoit, N.; Fall, M.; Nakoulima, A.; Dionne, P.; Fall, K.B.; Diatta, B.; et al. Emergence of Mutations in the K13 Propeller Gene of Plasmodium falciparum Isolates from Dakar, Senegal, in 2013–2014. Antimicrob. Agents Chemother. 2016, 60, 624–627. [Google Scholar] [CrossRef] [PubMed]
- Group, W.K.G.-P.S. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3d7_1343700) with parasite clearance rates after artemisinin-based treatments-a WWARN individual patient data meta-analysis. BMC Med. 2019, 17, 1. [Google Scholar]
- Wu, Y.; Soe, M.T.; Aung, P.L.; Zhao, L.; Zeng, W.; Menezes, L.; Yang, Z.; Kyaw, M.P.; Cui, L. Efficacy of artemether-lumefantrine for treating uncomplicated Plasmodium falciparum cases and molecular surveillance of drug resistance genes in Western Myanmar. Malar. J. 2020, 19, 304. [Google Scholar] [CrossRef]
- Ashley, E.A.; Dhorda, M.; Fairhurst, R.M.; Amaratunga, C.; Lim, P.; Suon, S.; Sreng, S.; Anderson, J.M.; Mao, S.; Sam, B.; et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014, 371, 411–423. [Google Scholar] [CrossRef]
- Takala-Harrison, S.; Jacob, C.G.; Arze, C.; Cummings, M.P.; Silva, J.C.; Dondorp, A.M.; Fukuda, M.M.; Hien, T.T.; Mayxay, M.; Noedl, H. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J. Infect. Dis. 2015, 211, 670–679. [Google Scholar] [CrossRef]
- Carrara, V.I.; Lwin, K.M.; Phyo, A.P.; Ashley, E.; Wiladphaingern, J.; Sriprawat, K.; Rijken, M.; Boel, M.; McGready, R.; Proux, S.; et al. Malaria burden and artemisinin resistance in the mobile and migrant population on the Thai-Myanmar border, 1999–2011: An observational study. PLoS Med. 2013, 10, e1001398. [Google Scholar] [CrossRef]
- Saunders, D.L.; Vanachayangkul, P.; Lon, C. Dihydroartemisinin-piperaquine failure in Cambodia. N. Engl. J. Med. 2014, 371, 484–485. [Google Scholar] [CrossRef] [PubMed]
- Spring, M.D.; Lin, J.T.; Manning, J.E.; Vanachayangkul, P.; Somethy, S.; Bun, R.; Se, Y.; Chann, S.; Ittiverakul, M.; Sia-ngam, P.; et al. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: An observational cohort study. Lancet Infect. Dis. 2015, 15, 683–691. [Google Scholar] [CrossRef] [PubMed]
- van der Pluijm, R.W.; Tripura, R.; Hoglund, R.M.; PyaePhyo, A.; Lek, D.; Ul Islam, A.; Anvikar, A.R.; Satpathi, P.; Satpathi, S.; Behera, P.K.; et al. Triple artemisinin-based combination therapies versus artemisinin-based combination therapies for uncomplicated Plasmodium falciparum malaria: A multicentre, open-label, randomised clinical trial. Lancet 2020, 395, 1345–1360. [Google Scholar] [CrossRef]
- Liu, H.; Yang, H.L.; Tang, L.H.; Li, X.L.; Huang, F.; Wang, J.Z.; Li, C.F.; Wang, H.Y.; Nie, R.H.; Guo, X.R.; et al. In vivo monitoring of dihydroartemisinin-piperaquine sensitivity in Plasmodium falciparum along the China-Myanmar border of Yunnan Province, China from 2007 to 2013. Malar. J. 2015, 14, 47. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Si, Y.; Zeng, W.; Li, N.; Wang, C.; Siddiqui, F.; Zhang, J.; Pi, L.; He, X.; Zhao, L.; Wang, S.; et al. In Vitro Susceptibility of Plasmodium falciparum Isolates from the China-Myanmar Border Area to Piperaquine and Association with Candidate Markers. Antimicrob. Agents Chemother. 2021, 65, e02305-20. [Google Scholar] [CrossRef]
- Witkowski, B.; Duru, V.; Khim, N.; Ross, L.S.; Saintpierre, B.; Beghain, J.; Chy, S.; Kim, S.; Ke, S.; Kloeung, N.; et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: A phenotype-genotype association study. Lancet Infect. Dis. 2017, 17, 174–183. [Google Scholar] [CrossRef]
- Amato, R.; Lim, P.; Miotto, O.; Amaratunga, C.; Dek, D.; Pearson, R.D.; Almagro-Garcia, J.; Neal, A.T.; Sreng, S.; Suon, S.; et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: A genotype-phenotype association study. Lancet Infect. Dis. 2017, 17, 164–173. [Google Scholar] [CrossRef]
- Dhingra, S.K.; Small-Saunders, J.L.; Menard, D.; Fidock, D.A. Plasmodium falciparum resistance to piperaquine driven by PfCRT. Lancet Infect. Dis. 2019, 19, 1168–1169. [Google Scholar] [CrossRef]
- Ross, L.S.; Dhingra, S.K.; Mok, S.; Yeo, T.; Wicht, K.J.; Kumpornsin, K.; Takala-Harrison, S.; Witkowski, B.; Fairhurst, R.M.; Ariey, F.; et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat. Commun. 2018, 9, 3314. [Google Scholar] [CrossRef]
- Duru, V.; Khim, N.; Leang, R.; Kim, S.; Domergue, A.; Kloeung, N.; Ke, S.; Chy, S.; Eam, R.; Khean, C.; et al. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: Retrospective and prospective investigations. BMC Med. 2015, 13, 305. [Google Scholar] [CrossRef]
- Miotto, O.; Amato, R.; Ashley, E.A.; MacInnis, B.; Almagro-Garcia, J.; Amaratunga, C.; Lim, P.; Mead, D.; Oyola, S.O.; Dhorda, M.; et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015, 47, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.P.; Dave, A.; Stein, W.D.; Lanzer, M. Transporters as mediators of drug resistance in Plasmodium falciparum. Int. J. Parasitol. 2010, 40, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Price, R.N.; Uhlemann, A.C.; Brockman, A.; McGready, R.; Ashley, E.; Phaipun, L.; Patel, R.; Laing, K.; Looareesuwan, S.; White, N.J.; et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 2004, 364, 438–447. [Google Scholar] [CrossRef] [PubMed]
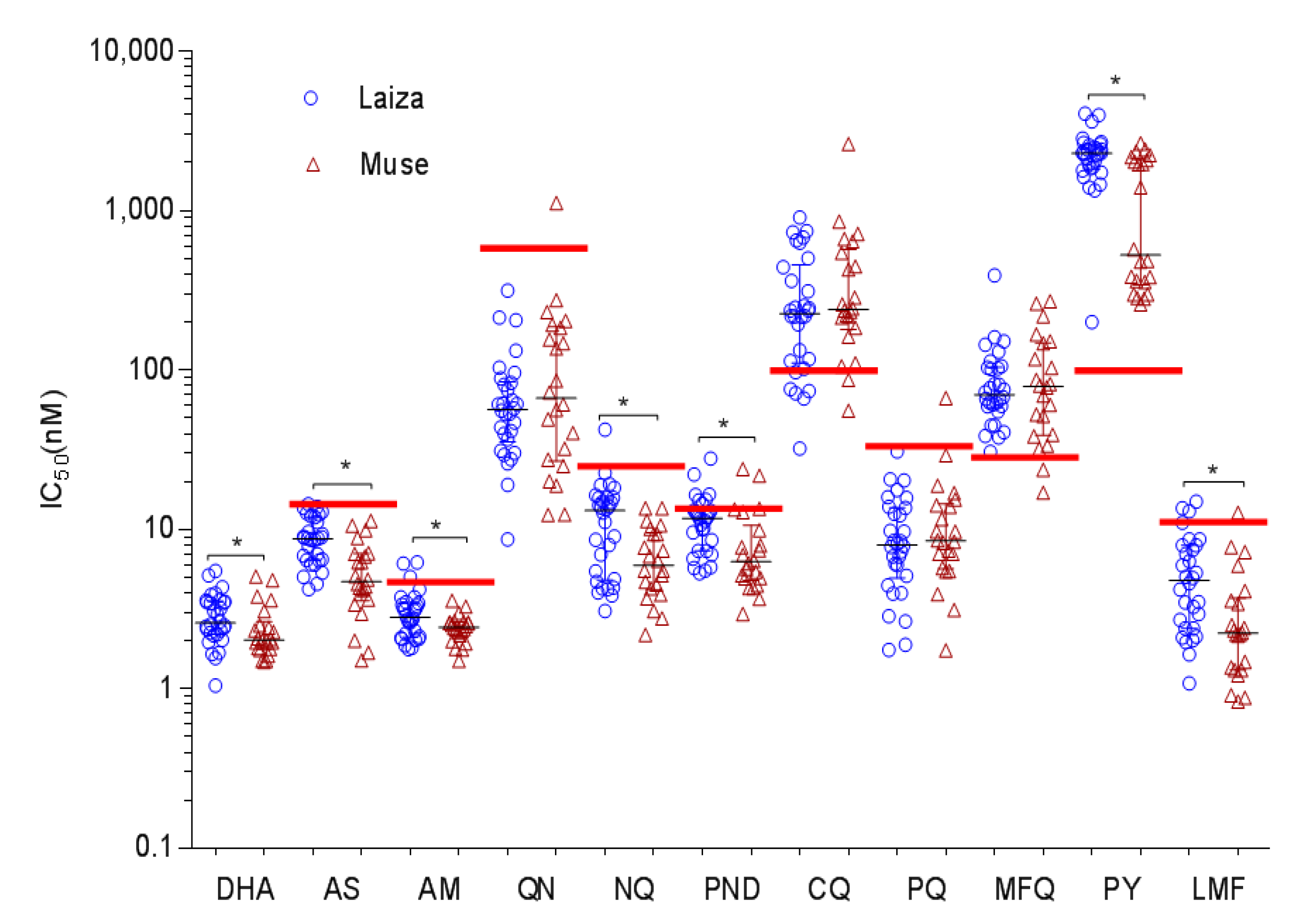
| Drug | Total (N = 52) (Mean ± SD) | 3D7 Control (Mean ± SD) | p-Value * Field Sites vs. Control | Laiza (N = 30) | Muse (N = 22) | p-Value # Laiza vs. Muse | ||
|---|---|---|---|---|---|---|---|---|
| (Mean ± SD) | Range | (Mean ± SD) | Range | Muse | ||||
| CQ | 358 ±394.9 | 17.98 ± 0.53 | <0.0001 | 303.3 ±241.6 | 32.14–903.23 | 432.6 ±536.7 | 55.41–2616 | 0.4010 |
| PPQ | 10.98 ±10.11 | 5.20 ± 0.26 | <0.0001 | 9.76 ±6.58 | 1.75–30.71 | 12.64 ±13.55 | 1.74–66.65 | 0.5847 |
| MFQ | 93.49 ±69.44 | 15.40 ± 0.37 | <0.0001 | 89.51 ±66.73 | 30.56–391.87 | 98.92 ±74.22 | 16.99–269.53 | 0.8869 |
| NQ | 9.93 ±6.94 | 5.36 ±2.39 | <0.0001 | 12.17 ±8.01 | 3.08–42.31 | 6.88 ±3.41 | 2.18–13.59 | 0.0053 |
| PY | 1796 ±989.4 | 66.73 ± 1.28 | <0.0001 | 2259 ±763.8 | 199.97–4058.0 | 1166 ±922 | 258.3–2640.0 | 0.0003 |
| AS | 7.46 ±3.34 | 5.43 ± 2.01 | 0.0002 | 8.92 ±2.98 | 4.22–14.37 | 5.48 ±2.78 | 1.51–11.32 | 0.0001 |
| DHA | 2.70 ±1.05 | 0.73 ± 0.13 | <0.0001 | 2.90 ±1.04 | 1.05–5.49 | 2.42 ±1.02 | 1.48–5.05 | 0.0319 |
| AM | 2.79 ±0.96 | 1.36 ± 1.59 | <0.0001 | 3.06 ±1.13 | 1.77–6.19 | 2.43 ±0.48 | 1.5–3.58 | 0.0244 |
| QN | 103.6 ±159.9 | 86.75 ± 1.91 | 0.2790 | 74.59 ±65.5 | 8.65–314.7 | 143.1 ±230.9 | 12.33–1113 | 0.5481 |
| LMF | 4.58 ±3.60 | 4.60 ± 2.90 | 0.4193 | 5.60 ±3.77 | 1.08–14.93 | 3.18 ±2.89 | 0.83–12.71 | 0.0038 |
| PND | 10.27 ±5.445 | 8.51 ± 1.38 | 0.0533 | 11.69 ±4.96 | 5.31–27.87 | 8.34 ±5.59 | 2.94–23.88 | 0.0031 |
| RSA a | 3.83% (1.83–7.26%) | 0.3% | <0.0001 | 4.99% (1.87–8.4) | 2.4% (1.72–3.79) | 0.0495 | ||
| Gene | Mutations | Total (n = 52) | Laiza (n = 30) | Muse (n = 22) | p-Value |
|---|---|---|---|---|---|
| Pfdhfr | N51I | 73.1 | 96.7 | 40.9 | <0.0001 * |
| C59R | 100 | 100 | 100 | 1.0000 | |
| S108N | 100 | 100 | 100 | 1.0000 | |
| I164L | 96.2 | 100 | 90.9 | 0.1742 | |
| Pfdhps | S436A | 48.1 | 33.3 | 68.2 | 0.0238 * |
| K540E/N | 96.2 | 96.7 | 95.5 | 1.0000 | |
| A581G | 55.8 | 73.3 | 31.8 | 0.0046 * | |
| Pfcrt | C72S | 19.2 | 6.7 | 36.4 | 0.0117 * |
| M74I | 100 | 100 | 100 | 1.0000 | |
| N75E | 100 | 100 | 100 | 1.0000 | |
| K76T | 100 | 100 | 100 | 1.0000 | |
| A220S | 98.1 | 100 | 95.5 | 0.4231 | |
| I356T | 100 | 100 | 100 | 1.0000 | |
| Pfmdr1 | N86Y | 1.9 | 3.3 | 0 | 1.0000 |
| G130K | 5.8 | 0 | 13.6 | 0.0697 | |
| Y184F | 40.4 | 70 | 0 | <0.0001 * | |
| S1034I | 1.9 | 0 | 4.5 | 0.4231 | |
| N1042D | 1.9 | 0 | 4.5 | 0.4231 | |
| F1226Y | 13.5 | 0 | 31.8 | 0.0013 * | |
| Pfmrp1 | H191Y | 90.4 | 86.7 | 95.5 | 0.3814 |
| N325S | 5.8 | 3.3 | 9.1 | 0.5670 | |
| S437A | 90.4 | 86.7 | 95.5 | 0.3814 | |
| H785N | 71.2 | 83.3 | 54.5 | 0.0324 * | |
| I876V | 76.9 | 80 | 72.7 | 0.7402 | |
| T1007M | 73.1 | 83.3 | 59.1 | 0.0647 | |
| F1390I | 5.8 | 0 | 13.6 | 1.0000 | |
| PfK13 | NN insert | 88.5 | 96.7 | 77.3 | 0.0716 |
| K189T | 3.8 | 0 | 9.1 | 0.1742 | |
| F446I | 63.5 | 86.7 | 31.8 | <0.0001 * | |
| N458Y | 3.8 | 0 | 9.1 | 0.1742 |
| Gene (Codon Positions) | Haplotypes | Total (n = 52) | Laiza (n = 30) | Muse (n = 22) | p-Value |
|---|---|---|---|---|---|
| Pfdhfr (51/59/108/164) | NRNI | 3.8 | 0 | 9.1 | 0.1742 |
| NRNL | 23.1 | 3.3 | 50.0 | 0.0001 * | |
| IRNL | 73.1 | 96.7 | 40.9 | <0.0001 * | |
| Pfdhps (436/540/581) | SKA | 1.9 | 0 | 4.5 | 0.4231 |
| SKG | 1.9 | 3.3 | 0 | 1.0000 | |
| SNG | 40.4 | 56.7 | 18.2 | 0.0093 * | |
| SEG | 7.7 | 6.7 | 9.1 | 1.0000 | |
| ANA | 3.8 | 0 | 9.1 | 0.1742 | |
| AEA | 38.5 | 26.7 | 54.5 | 0.0499 * | |
| AEG | 3.8 | 3.3 | 4.5 | 1.0000 | |
| ANG | 1.9 | 3.3 | 0 | 1.0000 | |
| Pfcrt (72/74/75/76/220) | CIETA | 1.9 | 0 | 4.5 | 0.4231 |
| CIETS | 78.8 | 93.3 | 59.1 | 0.0048 * | |
| SIETS | 19.2 | 6.7 | 36.4 | 0.0117 * | |
| Pfmdr1 (86/130/184/1042) | NEYSNF | 36.5 | 26.7 | 50.0 | 0.1442 |
| NEYSNY | 11.5 | 0 | 27.3 | 0.0037 * | |
| NEYINF | 1.9 | 0 | 4.5 | 0.4231 | |
| NEFSNF | 40.4 | 70.0 | 0 | <0.0001 * | |
| NKYSNF | 5.8 | 0 | 13.6 | 0.0697 | |
| YEYSNF | 1.9 | 3.3 | 0 | 1.0000 | |
| NEYSDY | 1.9 | 0 | 4.5 | 0.4231 | |
| Pfmrp1 (191/325/437/785/ 876/1007/1390) | HNSHITF | 9.6 | 13.3 | 4.5 | 0.3814 |
| YNAHITF | 5.8 | 0 | 13.6 | 0.0697 | |
| YSAHITF | 5.8 | 3.3 | 9.1 | 0.5670 | |
| YNAHVTI | 5.8 | 0 | 13.6 | 0.0697 | |
| YNAHVMF | 1.9 | 0 | 4.5 | 0.4231 | |
| YNANIMF | 1.9 | 3.3 | 0 | 1.0000 | |
| YNANVMF | 69.2 | 80.0 | 54.5 | 0.0701 | |
| PfK13 (189/446/458) | KFN | 28.8 | 13.3 | 50.0 | 0.0057 * |
| KIN | 63.5 | 86.7 | 31.8 | <0.0001 * | |
| KFY | 3.8 | 0 | 9.1 | 0.1742 | |
| TFN | 3.8 | 0 | 9.1 | 0.1742 | |
| Pfnhe1 | MS-3 | 3.8 | 0 | 9.1 | 0.1742 |
| MS-5 | 40.4 | 66.7 | 4.6 | <0.0001 * | |
| MS-6 | 17.3 | 26.7 | 4.6 | 0.0618 | |
| MS-7 | 34.6 | 6.7 | 72.7 | <0.0001 * | |
| MS-21 | 3.8 | 0 | 9.1 | 0.1742 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, M.; Bai, Y.; Deng, S.; Ruan, Y.; Zeng, W.; Li, X.; Wang, X.; Zhao, W.; Zhao, H.; Sun, K.; et al. Different In Vitro Drug Susceptibility Profile of Plasmodium falciparum Isolates from Two Adjacent Areas of Northeast Myanmar and Molecular Markers for Drug Resistance. Trop. Med. Infect. Dis. 2022, 7, 442. https://doi.org/10.3390/tropicalmed7120442
Duan M, Bai Y, Deng S, Ruan Y, Zeng W, Li X, Wang X, Zhao W, Zhao H, Sun K, et al. Different In Vitro Drug Susceptibility Profile of Plasmodium falciparum Isolates from Two Adjacent Areas of Northeast Myanmar and Molecular Markers for Drug Resistance. Tropical Medicine and Infectious Disease. 2022; 7(12):442. https://doi.org/10.3390/tropicalmed7120442
Chicago/Turabian StyleDuan, Mengxi, Yao Bai, Shuang Deng, Yonghua Ruan, Weilin Zeng, Xiaosong Li, Xun Wang, Wei Zhao, Hui Zhao, Kemin Sun, and et al. 2022. "Different In Vitro Drug Susceptibility Profile of Plasmodium falciparum Isolates from Two Adjacent Areas of Northeast Myanmar and Molecular Markers for Drug Resistance" Tropical Medicine and Infectious Disease 7, no. 12: 442. https://doi.org/10.3390/tropicalmed7120442
APA StyleDuan, M., Bai, Y., Deng, S., Ruan, Y., Zeng, W., Li, X., Wang, X., Zhao, W., Zhao, H., Sun, K., Zhu, W., Wu, Y., Miao, J., Kyaw, M. P., Yang, Z., & Cui, L. (2022). Different In Vitro Drug Susceptibility Profile of Plasmodium falciparum Isolates from Two Adjacent Areas of Northeast Myanmar and Molecular Markers for Drug Resistance. Tropical Medicine and Infectious Disease, 7(12), 442. https://doi.org/10.3390/tropicalmed7120442






