Analysis of a Dengue Virus Outbreak in Rosso, Senegal 2021
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
2.1.1. Syndromic Sentinel Surveillance Network in Senegal (4S Network)
2.1.2. Study Sites
2.2. Outbreak Investigation and Patient Enrolment
- -
- Suspected case: sudden onset of fever (>38 °C) in a resident of the survey area, with two or more of the following signs/symptoms: headache, retro-orbital pain, arthralgia, myalgia, nausea/vomiting, rash, hemorrhagic manifestations, leukopenia.
- -
- Probable confirmed case: Any suspected case with a positive Dengue RDT (IgM/IgG or detection of viral antigen NS1).
- -
- Confirmed case: Suspect or probable case confirmed by the reference laboratory (positive IgM serology, increase in IgG titers, virus detection by RT-PCR or isolation on cell culture).
2.3. NS1/IgM-IgG Rapid Detection Tests Detection on the Field
2.4. Molecular Diagnostics for DENV Detection and Serotyping
2.5. Serological Assays for Specific DENV Antibodies Detection
2.6. Sequencing and Phylogenetic Analysis
2.7. Statistical Analysis
2.8. Ethical Consideration
3. Results
3.1. Demographic Characteristics and Epidemiology
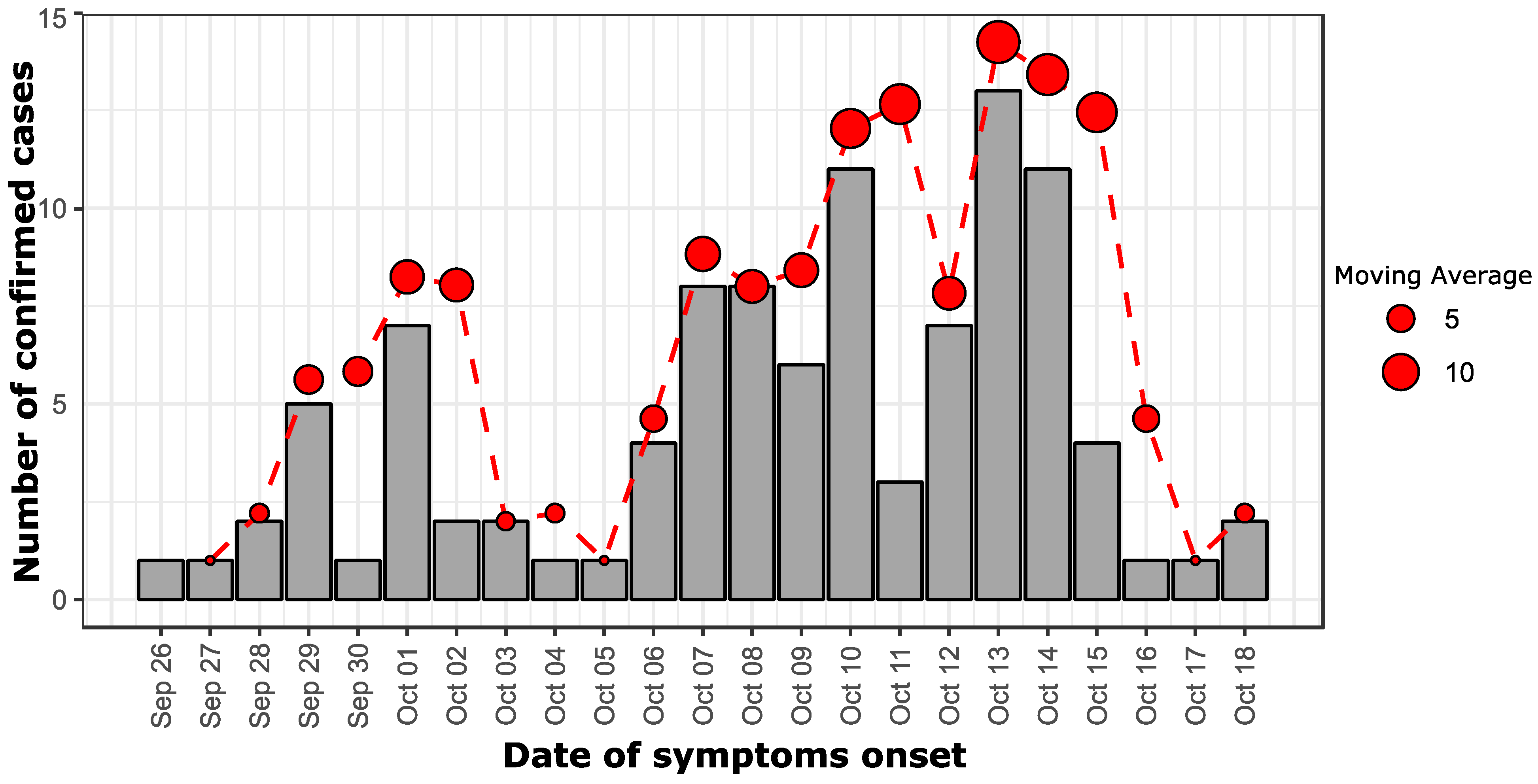
3.2. Epidemiological Curve
3.3. Circulating Dengue Virus Serotypes
3.4. Comparison of NS1 vs. qRT-PCR Performance
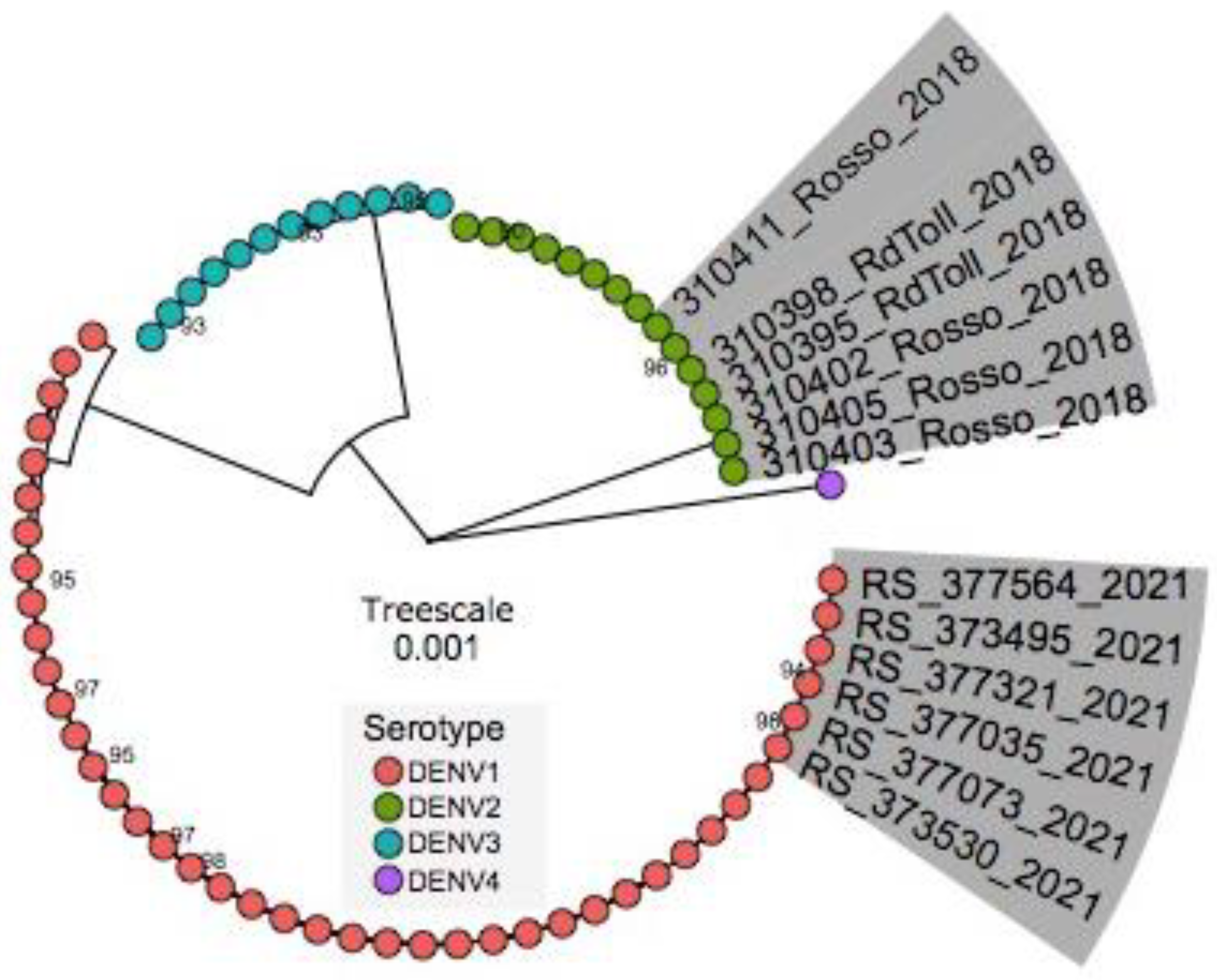
3.5. Phylogenetic Inference
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hotta, S. Experimental studies on dengue. I. Isolation, identification and modification of the virus. J. Infect. Dis. 1952, 90, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, M.U.G.; Sinka, M.E.; Duda, K.A.; Mylne, A.Q.N.; Shearer, F.M.; Barker, C.M.; Moore, C.G.; Carvalho, R.G.; Coelho, G.E.; Van Bortel, W.; et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 2015, 4, e08347. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Brady, O.J.; Golding, N.; Kraemer, M.U.G.; Wint, G.R.W.; Ray, S.E.; Pigott, D.M.; Shearer, F.M.; Johnson, K.; Earl, L.; et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019, 4, 1508–1515. [Google Scholar] [CrossRef]
- WHO/TDR (Ed.) Dengue: Guidelines for Diagnosis, Treatment, Prevention, and Control; New Edtion; TDR: World Health Organization: Geneva, Switzerland, 2009; 147p. [Google Scholar]
- WHO. Global Strategy for Dengue Prevention and Control, 2012–2020; World Health Organization: Geneva, Switzerland, 2012; Available online: http://apps.who.int/iris/bitstream/10665/75303/1/9789241504034_eng.pdf (accessed on 12 September 2020).
- Gubler, D.J. Dengue and Dengue Hemorrhagic Fever. Clin. Microbiol. Rev. 1998, 11, 480–496. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Were, F. The dengue situation in Africa. Paediatr. Int. Child Health 2012, 32, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, A.; Kuritsky, J.N.; Letson, G.W.; Margolis, H.S. Dengue Virus Infection in Africa. Emerg. Infect. Dis. 2011, 17, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Tarnagda, Z.; Cissé, A.; Bicaba, B.W.; Diagbouga, S.; Sagna, T.; Ilboudo, A.K.; Tialla, D.; Lingani, M.; Sondo, K.A.; Yougbaré, I.; et al. Dengue Fever in Burkina Faso, 2016. Emerg. Infect. Dis. 2018, 24, 170–172. [Google Scholar] [CrossRef]
- WHO. Regional Office for Africa Weekly Bulletin on Outbreaks and Other Emergences 2017. Available online: https://reliefweb.int/sites/reliefweb.int/files/resources/OEW27-1772017.pdf (accessed on 27 October 2018).
- Gaye, A.; Wang, E.; Vasilakis, N.; Guzman, H.; Diallo, D.; Talla, C.; Ba, Y.; Dia, I.; Weaver, S.C.; Diallo, M. Potential for sylvatic and urban Aedes mosquitoes from Senegal to transmit the new emerging dengue serotypes 1, 3 and 4 in West Africa. PLoS Negl. Trop. Dis. 2019, 13, e0007043. [Google Scholar] [CrossRef]
- Robin, Y.; Cornet, M.; Heme, G.; Le Gonidec, G. Isolement du virus de la dengue au Sénégal. Annales de l’Institut Pasteur/Virologie 1980, 131, 149–154. [Google Scholar] [CrossRef]
- Zeller, H.; Cornet, J.; Diop, A.; Camicas, J. Crimean-Congo hemorrhagic fever in ticks (Acari: Ixodidae) and ruminants: Field observations of an epizootic in Bandia, Sénégal. J. Med. Entomol. 1997, 4, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Faye, O.; Ba, Y.; Faye, O.; Talla, C.; Diallo, D.; Chen, R.; Mondo, M.; Ba, R.; Macondo, E.; Siby, T.; et al. Urban Epidemic of Dengue Virus Serotype 3 Infection, Senegal, 2009. Emerg. Infect. Dis. 2014, 20, 456–459. [Google Scholar] [CrossRef] [PubMed]
- Dieng, I.; Ndione, M.H.D.; Fall, C.; Diagne, M.M.; Diop, M.; Gaye, A.; Barry, M.A.; Diop, B.; Ndiaye, M.; Bousso, A.; et al. Multifoci and multiserotypes circulation of dengue virus in Senegal between 2017 and 2018. BMC Infect. Dis. 2021, 21, 867. [Google Scholar] [CrossRef] [PubMed]
- Diallo, M.; Ba, Y.; Sall, A.A.; Diop, O.M.; Ndione, J.A.; Mondo, M.; Girault, L.; Mathiot, C. Amplification of the Sylvatic Cycle of Dengue Virus Type 2, Senegal, 1999–2000: Entomologic Findings and Epidemiologic Considerations. Emerg. Infect. Dis. 2003, 9, 362–367. [Google Scholar] [CrossRef]
- Dieng, I.; Hedible, B.G.; Diagne, M.M.; El Wahed, A.A.; Diagne, C.T.; Fall, C.; Richard, V.; Vray, M.; Weidmann, M.; Faye, O.; et al. Mobile Laboratory Reveals the Circulation of Dengue Virus Serotype I of Asian Origin in Medina Gounass (Guediawaye), Senegal. Diagnostics 2020, 10, 408. [Google Scholar] [CrossRef]
- Gaye, A.; Ndiaye, T.; Sy, M.; Deme, A.B.; Thiaw, A.B.; Sene, A.; Ndiaye, C.; Diedhiou, Y.; Mbaye, A.M.; Ndiaye, I.; et al. Genomic investigation of a dengue virus outbreak in Thiès, Senegal, in 2018. Sci. Rep. 2021, 11, 10321. [Google Scholar] [CrossRef]
- Diagne, C.T.; Barry, M.A.; Ba, Y.; Faye, O.; Sall, A.A. Dengue epidemic in Touba, Senegal: Implications for the Grand Magal Pilgrimage for travellers. J. Travel Med. 2019, 26, tay123. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Calisher, C.H.; Gubler, D.J.; Chang, G.J.; Vorndam, A.V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 545–551. [Google Scholar] [CrossRef]
- Seah, C.L.K.; Chow, V.T.K.; Tan, H.C.; Chan, Y.C. Rapid, single-step RT-PCR typing of dengue viruses using five NS3 gene primers. J. Virol. Methods 1995, 51, 193–200. [Google Scholar] [CrossRef]
- Laue, T.; Emmerich, P.; Schmitz, H. Detection of Dengue Virus RNA in Patients after Primary or Secondary Dengue Infection by Using the TaqMan Automated Amplification System. J. Clin. Microbiol. 1999, 37, 2543–2547. [Google Scholar] [CrossRef]
- Oyero, O.G.; Ayukekbong, J.A. High dengue NS1 antigenemia in febrile patients in Ibadan, Nigeria. Virus Res. 2014, 191, 59–61. [Google Scholar] [CrossRef]
- Aryati, A.; Trimarsanto, H.; Yohan, B.; Wardhani, P.; Fahri, S.; Sasmono, R.T. Performance of commercial dengue NS1 ELISA and molecular analysis of NS1 gene of dengue viruses obtained during surveillance in Indonesia. BMC Infect. Dis. 2013, 13, 611. [Google Scholar] [CrossRef] [PubMed]
- Dia, N.; Diene Sarr, F.; Thiam, D.; Faye Sarr, T.; Espié, E.; OmarBa, I.; Coly, M.; Niang, M.; Richard, V. Influenza-Like Illnesses in Senegal: Not Only Focus on Influenza Viruses. PLoS ONE 2014, 9, e93227. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3968133/ (accessed on 24 October 2019). [CrossRef] [PubMed]
- Yow, K.-S.; Aik, J.; Tan, E.Y.-M.; Ng, L.-C.; Lai, Y.-L. Rapid diagnostic tests for the detection of recent dengue infections: An evaluation of six kits on clinical specimens. PLoS ONE 2021, 16, e0249602. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vargas, L.A.; Sánchez-Marce, E.E.; Vivanco-Cid, H. Evaluation of the SD BIOLINE Dengue Duo rapid test in the course of acute and convalescent dengue infections in a Mexican endemic region. Diagn. Microbiol. Infect. Dis. 2014, 78, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; de With, K.; Huzly, D.; Hufert, F.; Weidmann, M.; Breisinger, S.; Eppinger, S.; Kern, W.V.; Bauer, T.M. Nosocomial Acquisition of Dengue. Emerg. Infect. Dis. 2004, 10, 1872–1873. [Google Scholar] [CrossRef] [PubMed]
- Dieng, I.; dos Passos Cunha, M.; Diagne, M.M.; Sembène, P.M.; de Andrade Zanotto, P.M.; Faye, O.; Faye, O.; Sall, A.A. Origin and Spread of the Dengue Virus Type 1, Genotype V in Senegal, 2015–2019. Viruses 2021, 13, 57. [Google Scholar] [CrossRef]
- Ba, F.; Loucoubar, C.; Faye, O.; Fall, G.; Noelle Mbaye, R.N.P.; Sembene, M.; Diallo, M.; Toure Balde, A.; Alpha Sall, A.; Faye, O. Retrospective analysis of febrile patients reveals unnoticed epidemic of zika fever in Dielmo, Senegal, 2000. Clin. Microbiol. Infect. Dis. 2018, 3, 1–9. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Nguyen, T.H.T.; Clapham, H.E.; Phung, K.L.; Nguyen, T.K.; DInh, T.T.; Nguyen, T.H.Q.; Tran, V.N.; Whitehead, S.; Simmons, C.; Wolbers, M.; et al. Methods to discriminate primary from secondary dengue during acute symptomatic infection. BMC Infect. Dis. 2018, 18, 375. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Dieng, I.; Diarra, M.; Diagne, M.M.; Faye, M.; Dior Ndione, M.H.; Ba, Y.; Diop, M.; Ndiaye, E.H.; Marinho de Andrade Zanotto, P.; Diop, B.; et al. Field Deployment of a Mobile Biosafety Laboratory Reveals the Co-Circulation of Dengue Viruses Serotype 1 and Serotype 2 in Louga City, Senegal, 2017. J. Trop. Med. 2021, 2021, 8817987. [Google Scholar] [CrossRef] [PubMed]
- Reich, N.G.; Shrestha, S.; King, A.A.; Rohani, P.; Lessler, J.; Kalayanarooj, S.; Yoon, I.-K.; Gibbons, R.V.; Burke, D.S.; Cummings, D.A.T. Interactions between serotypes of dengue highlight epidemiological impact of cross-immunity. J. R. Soc. Interface 2013, 10, 20130414. [Google Scholar] [CrossRef]
- Uno, N.; Ross, T.M. Dengue virus and the host innate immune response. Emerg. Microbes Infect. 2018, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Harapan, H.; Michie, A.; Sasmono, R.T.; Imrie, A. Dengue: A Minireview. Viruses 2020, 12, 829. [Google Scholar] [CrossRef] [PubMed]
- Simo Tchetgna, H.; Sado Yousseu, F.; Kamgang, B.; Tedjou, A.; McCall, P.J.; Wondji, C.S. Concurrent circulation of dengue serotype 1, 2 and 3 among acute febrile patients in Cameroon. PLoS Negl. Trop. Dis. 2021, 15, e0009860. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lormeau, V.-M.; Roche, C.; Aubry, M.; Teissier, A.; Lastere, S.; Daudens, E.; Mallet, H.-P.; Musso, D.; Aaskov, J. Recent emergence of dengue virus serotype 4 in French Polynesia results from multiple introductions from other South Pacific Islands. PLoS ONE 2011, 6, e29555. [Google Scholar] [CrossRef]
- Shrivastava, S.; Tiraki, D.; Diwan, A.; Lalwani, S.K.; Modak, M.; Mishra, A.C.; Arankalle, V.A. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS ONE 2018, 13, e0192672. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Fonville, J.M.; Gromowski, G.D.; Arriaga, J.B.; Green, A.; James, S.L.; Lau, L.; Montoya, M.; Wang, C.; VanBlargan, L.A.; et al. Dengue viruses cluster antigenically but not as discrete serotypes. Science 2015, 349, 1338–1343. [Google Scholar] [CrossRef]
- Shrivastava, A.; Soni, M.; Shrivastava, S.; Sharma, S.; Dash, P.K.; Gopalan, N.; Behera, P.K.; Parida, M.M. Lineage shift of dengue virus in Eastern India: An increased implication for DHF/DSS. Epidemiol. Infect. 2015, 143, 1599–1605. [Google Scholar] [CrossRef] [PubMed]
- Vilcarromero, S.; Chen, R.; Suarez-Ognio, L.; Johnson, W.L.; Morrison, A.C.; Long, K.C.; Wood, T.G.; Volkova, E.; Mayer, S.V.; Halsey, E.S.; et al. Lineage II of Southeast Asian/American DENV-2 is Associated with a Severe Dengue Outbreak in the Peruvian Amazon. Am. J. Trop. Med. Hyg. 2014, 91, 611–620. [Google Scholar] [CrossRef]
- Rico-Hesse, R.; Harrison, L.M.; Salas, R.A.; Tovar, D.; Nisalak, A.; Ramos, C.; Boshell, J.; de Mesa, M.T.R.; Nogueira, R.M.R.; Rosa, A.T. da Origins of Dengue Type 2 Viruses Associated with Increased Pathogenicity in the Americas. Virology 1997, 230, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Halstead, S.B. Immune enhancement of viral infection. Prog. Allergy 1982, 31, 301–364. [Google Scholar] [PubMed]
- Prajapati, S.; Napit, R.; Bastola, A.; Rauniyar, R.; Shrestha, S.; Lamsal, M.; Adhikari, A.; Bhandari, P.; Yadav, S.R.; Manandhar, K.D. Molecular phylogeny and distribution of dengue virus serotypes circulating in Nepal in 2017. PLoS ONE 2020, 15, e0234929. [Google Scholar] [CrossRef] [PubMed]
- Fourié, T.; El Bara, A.; Dubot-Pérès, A.; Grard, G.; Briolant, S.; Basco, L.K.; Ouldabdallahi Moukah, M.; Leparc-Goffart, I. Emergence of dengue virus serotype 2 in Mauritania and molecular characterization of its circulation in West Africa. PLoS Negl. Trop. Dis. 2021, 15, e0009829. [Google Scholar] [CrossRef]
- Louis, V.R.; Montenegro Quiñonez, C.A.; Kusumawathie, P.; Palihawadana, P.; Janaki, S.; Tozan, Y.; Wijemuni, R.; Wilder-Smith, A.; Tissera, H.A. Characteristics of and factors associated with dengue vector breeding sites in the City of Colombo, Sri Lanka. Pathog. Glob. Health 2016, 110, 79–86. [Google Scholar] [CrossRef]
- Anand, A.M. Evaluation of NS1 Antigen Detection for Early Diagnosis of Dengue in a Tertiary Hospital in Southern India. J. Clin. Diagn. Res. JCDR 2016, 10, DC01–DC04. [Google Scholar] [CrossRef]
- Amorim, J.H.; dos Santos Alves, R.P.; Boscardin, S.B.; de Souza Ferreira, L.C. The dengue virus non-structural 1 protein: Risks and benefits. Virus Res. 2014, 181, 53–60. [Google Scholar] [CrossRef]
- Hunsperger, E.A.; Yoksan, S.; Buchy, P.; Nguyen, V.C.; Sekaran, S.D.; Enria, D.A.; Vazquez, S.; Cartozian, E.; Pelegrino, J.L.; Artsob, H.; et al. Evaluation of Commercially Available Diagnostic Tests for the Detection of Dengue Virus NS1 Antigen and Anti-Dengue Virus IgM Antibody. PLoS Negl. Trop. Dis. 2014, 8, e3171. [Google Scholar] [CrossRef]
- Anker, M.; Arima, Y. Male–female differences in the number of reported incident dengue fever cases in six Asian countries. West. Pac. Surveill. Response J. WPSAR 2011, 2, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Yew, Y.W.; Ye, T.; Ang, L.W.; Ng, L.C.; Yap, G.; James, L.; Chew, S.K.; Goh, K.T. Seroepidemiology of dengue virus infection among adults in Singapore. Ann. Acad. Med. Singap. 2009, 38, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.; Celine, T. A descriptive study on dengue fever reported in a Medical College Hospital. Sahel Med. J. 2014, 17, 83–86. [Google Scholar] [CrossRef]
- Ooi, E.E. Changing Pattern of Dengue Transmission in Singapore. Chang. Pattern Dengue Transm. Singap. 2001, 25, 5. [Google Scholar]

| Total (n = 173) | Positives (n = 102) | Negatives (n = 71) | p_Value | |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| Median (IQR 1) | 21 ± (11–37) | 21 ± (9–36) | 23 ± (11–39) | 0.49 a |
| Age group (years) <15 15–30 30–45 >=45 | 64 (37.0) 51 (29.5) 25 (14.5) 33 (19.1) | 39 (38.2) 30 (29.4) 14 (14.0) 19 (18.6) | 25 (35.2) 21(30.0) 11 (15.5) 14 (20.0) | 0.97 b |
| Sex Female Male | 89 (51.4) 84 (48.6) | 46 (45.1) 56 (54.9) | 43 (60.6) 28 (39.4) | 0.04 b |
| Headache Yes No | 128 (74.0) 45 (26.0) | 82 (80.4) 20 (19.6) | 46 (65.0) 25 (35.2) | 0.02 b |
| Myalgia Yes No | 101 (59.0) 70 (41.0) | 64 (63.4) 37 (36.6) | 37 (53.0) 33 (69.0) | 0.17 b |
| Arthralgia Yes No | 107 (62.0) 66 (38.0) | 69 (67.6) 33 (32.4) | 38 (53.5) 33 (46.5) | 0.05 b |
| Asthenia Yes No | 64 (37.0) 109 (63.0) | 37 (36.3) 65 (63.7) | 27 (38.0) 44 (62.0) | 0.81 b |
| Abdominal pain Yes No | 8 (4.6) 165 (95.4) | 4 (4.0) 98(96.0) | 4 (5.6) 67 (94.4) | 0.60 b |
| Retro-orbital pain Yes No | 3 (2.0) 170 (98.0) | 2 (2.0) 100 (98.0) | 1 (1.4) 70 (98.6) | 0.63 b |
| Investigated health structures Rosso 1 (4S site) Mbagam PHC # Richard Toll DHS” Rosso 2 PHC # | 99 (57.2) 55 (32.0) 10 (6.0) 9 (5.2) | 73 (71.6) 21 (20.6) 2 (2.0) 6 (6.0) | 26 (36.6) 34 (48.0) 8 (11.3) 3 (4.2) | <0.001 b |
| Molecular Diagnostic | n (%) | 95 % CI |
|---|---|---|
| qRT-PCR all DENV | 64/176 (36, 36%) | 29.25–43.47 |
| DENV-1 | 50/64 (78, 12%) | 67.99–88.25 |
| DENV-2 | 00/64 | NA |
| DENV-3 | 00/64 | NA |
| DENV-4 | 00/64 | NA |
| Co-infections | 00/64 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dieng, I.; Barry, M.A.; Talla, C.; Sow, B.; Faye, O.; Diagne, M.M.; Sene, O.; Ndiaye, O.; Diop, B.; Diagne, C.T.; et al. Analysis of a Dengue Virus Outbreak in Rosso, Senegal 2021. Trop. Med. Infect. Dis. 2022, 7, 420. https://doi.org/10.3390/tropicalmed7120420
Dieng I, Barry MA, Talla C, Sow B, Faye O, Diagne MM, Sene O, Ndiaye O, Diop B, Diagne CT, et al. Analysis of a Dengue Virus Outbreak in Rosso, Senegal 2021. Tropical Medicine and Infectious Disease. 2022; 7(12):420. https://doi.org/10.3390/tropicalmed7120420
Chicago/Turabian StyleDieng, Idrissa, Mamadou Aliou Barry, Cheikh Talla, Bocar Sow, Oumar Faye, Moussa Moise Diagne, Ousseynou Sene, Oumar Ndiaye, Boly Diop, Cheikh Tidiane Diagne, and et al. 2022. "Analysis of a Dengue Virus Outbreak in Rosso, Senegal 2021" Tropical Medicine and Infectious Disease 7, no. 12: 420. https://doi.org/10.3390/tropicalmed7120420
APA StyleDieng, I., Barry, M. A., Talla, C., Sow, B., Faye, O., Diagne, M. M., Sene, O., Ndiaye, O., Diop, B., Diagne, C. T., Fall, G., Sall, A. A., Loucoubar, C., & Faye, O. (2022). Analysis of a Dengue Virus Outbreak in Rosso, Senegal 2021. Tropical Medicine and Infectious Disease, 7(12), 420. https://doi.org/10.3390/tropicalmed7120420








