Low Prevalence of Leptospira Carriage in Rodents in Leptospirosis-Endemic Northeastern Thailand
Abstract
1. Introduction
2. Materials and Methods
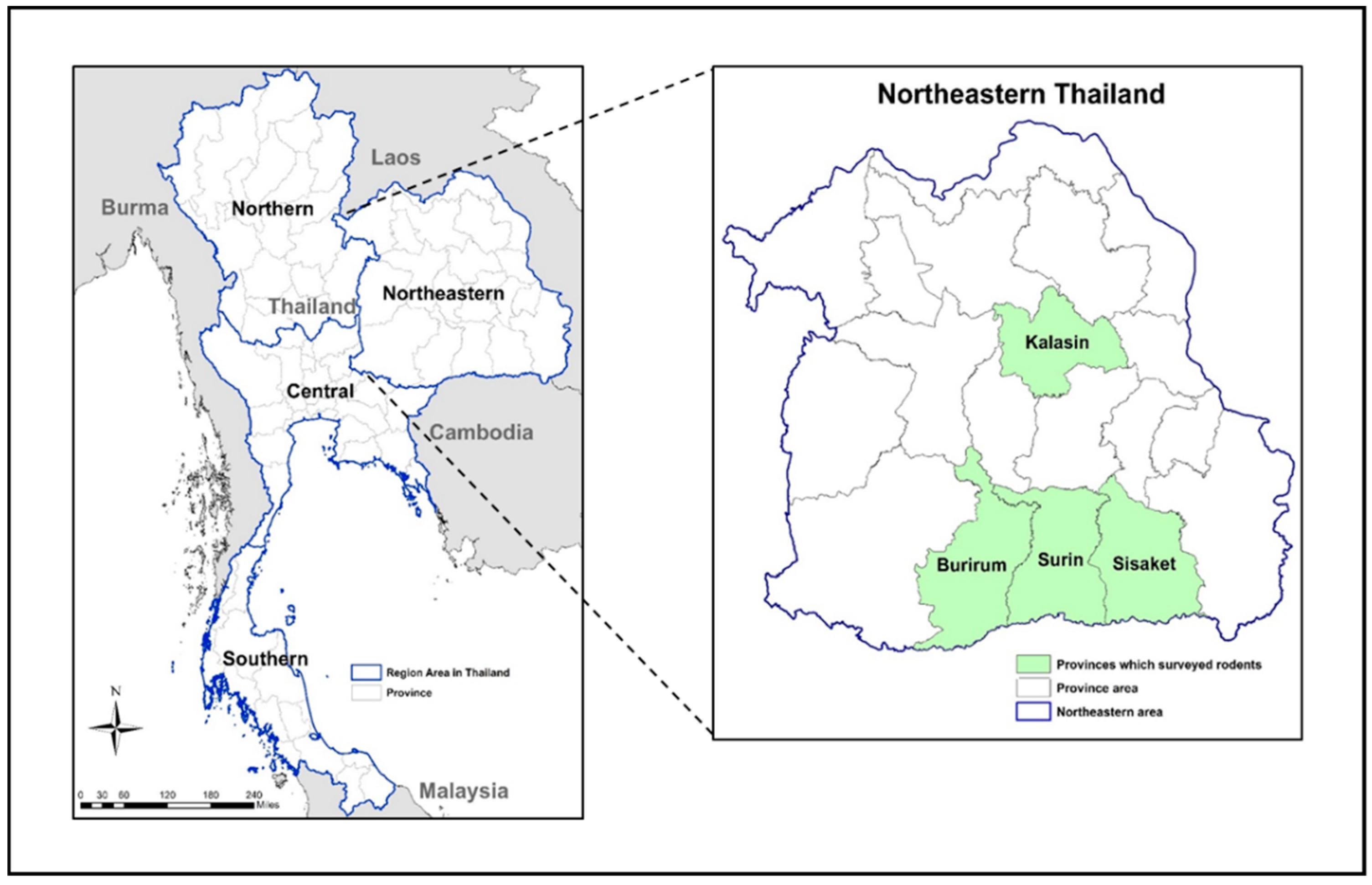
2.1. Trapping Rodents
2.2. Rodent Identification and Processing
2.3. Leptospira Cultures
2.4. Leptospira Carriage and Molecular Identification in Trapped Rodents
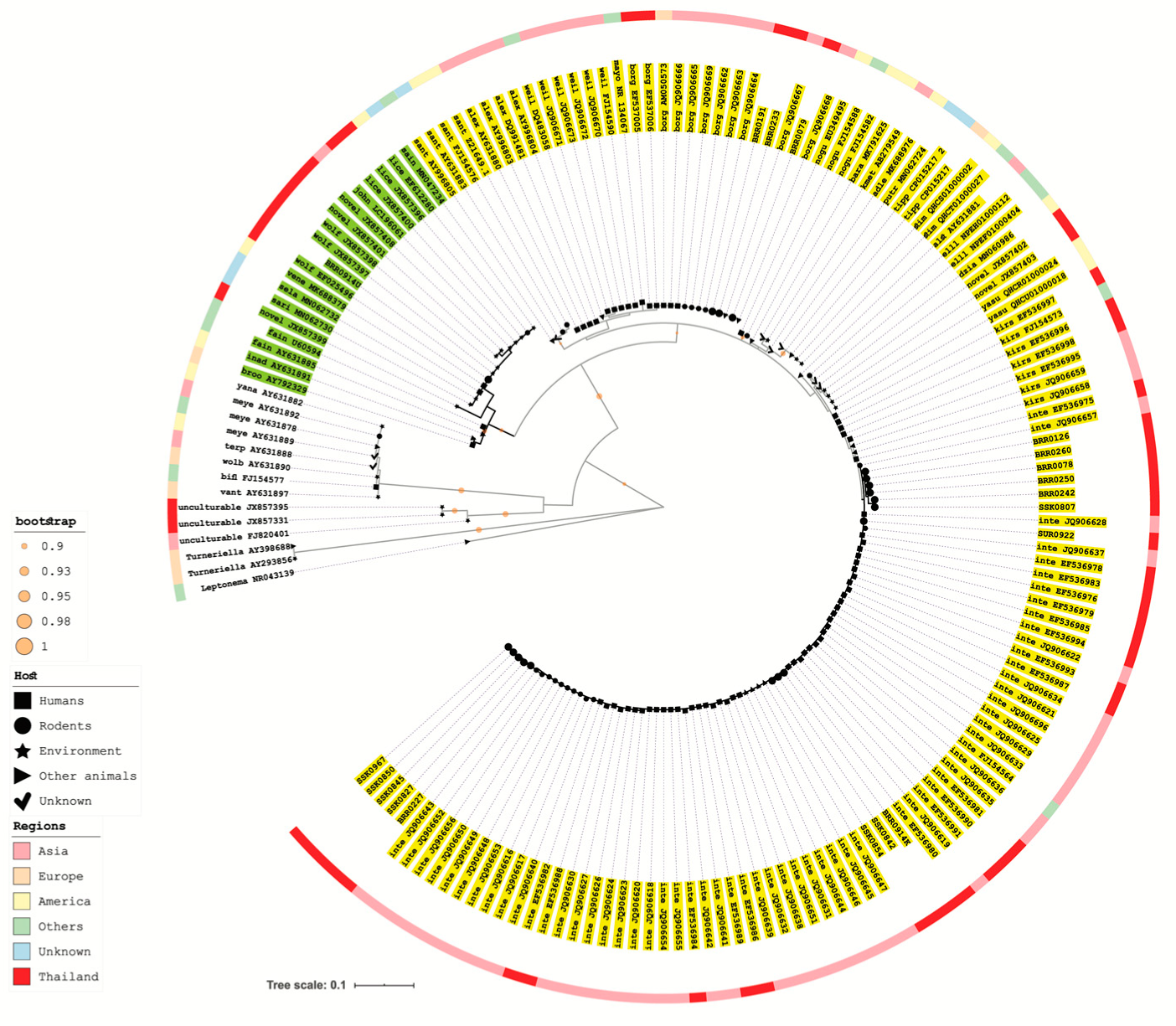
2.5. 16S rRNA Gene Amplification and Sequencing Analysis of Recovered Leptospira Isolate
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Faine, S.; Adler, B.; Bolin, C.; Perolat, P. “Leptospira” and Leptospirosis; Medisci Press: Melbourne, Australia, 1999. [Google Scholar]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef] [PubMed]
- Turk, N.; Milas, Z.; Margaletic, J.; Staresina, V.; Slavica, A.; Riquelme-Sertour, N.; Bellenger, E.; Baranton, G.; Postic, D. Molecular characterization of Leptospira spp. strains isolated from small rodents in Croatia. Epidemiol. Infect. 2003, 130, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771. [Google Scholar] [CrossRef]
- Ko, A.I.; Goarant, C.; Picardeau, M. Leptospira: The dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 2009, 7, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Tangkanakul, W.; Smits, H.L.; Jatanasen, S.; Ashford, D.A. Leptospirosis: An emerging health problem in Thailand. Southeast Asian J. Trop. Med. Public Health 2005, 36, 281–288. [Google Scholar] [PubMed]
- Wangroongsarb, P.; Petkanchanapong, W.; Yasaeng, S.; Imvithaya, A.; Naigowit, P. Survey of leptospirosis among rodents in epidemic areas of Thailand. J. Trop. Med. Parasitol. 2002, 25, 55–58. [Google Scholar]
- Leelarasamee, A.; Chupaprawan, C.; Chenchittikul, M.; Udompanthurat, S. Etiologies of acute undifferentiated febrile illness in Thailand. J. Med. Assoc. Thai 2004, 87, 464–472. [Google Scholar]
- Suttinont, C.; Losuwanaluk, K.; Niwatayakul, K.; Hoontrakul, S.; Intaranongpai, W.; Silpasakorn, S.; Suwancharoen, D.; Panlar, P.; Saisongkorh, W.; Rolain, J.M.; et al. Causes of acute, undifferentiated, febrile illness in rural Thailand: Results of a prospective observational study. Ann. Trop. Med. Parasitol. 2006, 100, 363–370. [Google Scholar] [CrossRef]
- Sejvar, J.; Tangkanakul, W.; Ratanasang, P.; Dowell, S.F.; Sangjun, N.; Bragg, S.; Ashford, D.; Tappero, J. An outbreak of leptospirosis, Thailand--the importance of the laboratory. Southeast Asian J. Trop. Med. Public Health 2005, 36, 289–295. [Google Scholar]
- National Disease Surveillance (Report 506). Thailand: Bureau of Epidemiology, Department of Disease Control, MoPH, Thailand. Available online: http://www.boe.moph.go.th/boedb/surdata/disease.php?dcontent=situation&ds=43 (accessed on 8 February 2019).
- Phulsuksombati, D.; Sangjun, N.; Khoprasert, Y.; Kingnate, D.; Tangkanakul, W. Leptospires in rodent, northeastern region 1999–2000. J. Health Sci. 2001, 10, 508–515. [Google Scholar]
- Nelson, L.; Clark, F.W. Correction for sprung traps in catch/effort calculations of trapping results. J. Mammal. 1973, 54, 295–298. [Google Scholar] [CrossRef]
- Lekagul, B.; McNeely, J.A. Mammals of Thailand; Association for the Conservation of Wildlife: Bangkok, Thailand, 1977. [Google Scholar]
- McAvin, J.C.; Kengluecha, A.; Takhampunya, R.; Richardson, J.H. A field-expedient method for detection of leptospirosis causative agents in rodents. U.S. Army Med Dep. J. 2012, 22–28. [Google Scholar]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Chierakul, W.; Limmathurotsakul, D.; Day, N.P.; Peacock, S.J. Molecular detection and speciation of pathogenic Leptospira spp. in blood from patients with culture-negative leptospirosis. BMC Infect. Dis. 2011, 11, 338. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Matthias, M.A.; Díaz, M.M.; Campos, K.J.; Calderon, M.; Willig, M.R.; Pacheco, V.; Gotuzzo, E.; Gilman, R.H.; Vinetz, J.M. Diversity of bat-associated Leptospira in the Peruvian Amazon inferred by bayesian phylogenetic analysis of 16S ribosomal DNA sequences. Am. J. Trop. Med. Hyg. 2005, 73, 964–974. [Google Scholar] [CrossRef]
- Cosson, J.F.; Picardeau, M.; Mielcarek, M.; Tatard, C.; Chaval, Y.; Suputtamongkol, Y.; Buchy, P.; Jittapalapong, S.; Herbreteau, V.; Morand, S. Epidemiology of Leptospira transmitted by rodents in Southeast Asia. PLoS Negl. Trop. Dis. 2014, 8, e2902. [Google Scholar] [CrossRef]
- Della Rossa, P.; Tantrakarnapa, K.; Sutdan, D.; Kasetsinsombat, K.; Cosson, J.F.; Supputamongkol, Y.; Chaisiri, K.; Tran, A.; Supputamongkol, S.; Binot, A.; et al. Environmental factors and public health policy associated with human and rodent infection by leptospirosis: A land cover-based study in Nan province, Thailand. Epidemiol. Infect. 2016, 144, 1550–1562. [Google Scholar] [CrossRef]
- Herbreteau, V.; Bordes, F.; Jittapalapong, S.; Supputamongkol, Y.; Morand, S. Rodent-borne diseases in Thailand: Targeting rodent carriers and risky habitats. Infect. Ecol. Epidemiol. 2012, 2, 18637. [Google Scholar] [CrossRef]
- Ivanova, S.; Herbreteau, V.; Blasdell, K.; Chaval, Y.; Buchy, P.; Guillard, B.; Morand, S. Leptospira and rodents in Cambodia: Environmental determinants of infection. Am. J. Trop. Med. Hyg. 2012, 86, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Thaipadungpanit, J.; Wuthiekanun, V.; Chierakul, W.; Smythe, L.D.; Petkanchanapong, W.; Limpaiboon, R.; Apiwatanaporn, A.; Slack, A.T.; Suputtamongkol, Y.; White, N.J.; et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl. Trop. Dis. 2007, 1, e56. [Google Scholar] [CrossRef] [PubMed]
- Herbreteau, V.; Gonzalez, J.P.; Andrianasolo, H.; Kittayapong, P.; Hugot, J.P. Mapping the potential distribution of Bandicota indica, vector of zoonoses in Thailand, by use of remote sensing and geographic information systems (a case of Nakhon Pathom province). Trop. Nat. Hist. 2005, 5, 61–67. [Google Scholar]
- Thitipramote, N.; Suwanjarat, J.; Breed, W.G. Reproductive biology of the greater bandicoot rat Bandicota indica (Rodentia: Muridae) in the rice fields of southern Thailand. Curr. Zool. 2009, 55, 48–55. [Google Scholar] [CrossRef]
- Lall, C.; Kumar, K.V.; Raj, R.V.; Vedhagiri, K.; Vijayachari, P. Prevalence and diversity of leptospires in different ecological niches of urban and rural areas of South Andaman Island. Microbes Environ. 2016, 31, 79–82. [Google Scholar] [CrossRef]
- Muñoz-Zanzi, C.; Mason, M.R.; Encina, C.; Astroza, A.; Romero, A. Leptospira contamination in household and environmental water in rural communities in southern Chile. Int. J. Environ. Res. Public Health 2014, 11, 6666–6680. [Google Scholar] [CrossRef]
- Bulach, D.M.; Zuerner, R.L.; Wilson, P.; Seemann, T.; McGrath, A.; Cullen, P.A.; Davis, J.; Johnson, M.; Kuczek, E.; Alt, D.P.; et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. USA 2006, 103, 14560–14565. [Google Scholar] [CrossRef]
- Monahan, A.M.; Callanan, J.J.; Nally, J.E. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect. Immun. 2008, 76, 4952–4958. [Google Scholar] [CrossRef]
- Thaipadungpanit, J.; Wuthiekanun, V.; Chantratita, N.; Yimsamran, S.; Amornchai, P.; Boonsilp, S.; Maneeboonyang, W.; Tharnpoophasiam, P.; Saiprom, N.; Mahakunkijcharoen, Y.; et al. Leptospira species in floodwater during the 2011 floods in the Bangkok Metropolitan Region, Thailand. Am. J. Trop. Med. Hyg. 2013, 89, 794–796. [Google Scholar] [CrossRef]
- Chaiwattanarungruengpaisan, S.; Suwanpakdee, S.; Sangkachai, N.; Chamsai, T.; Taruyanon, K.; Thongdee, M. Potentially pathogenic Leptospira species isolated from a waterfall in Thailand. Jpn. J. Infect. Dis. 2018, 71, 65–67. [Google Scholar] [CrossRef]
- Thongdee, M.; Chaiwattanarungruengpaisan, S.; Lekcharoen, P.; Yimchoho, N.; Buathong, R.; Wiriyarat, W. A Novel Genotype of Leptospira interrogans Recovered from Leptospirosis Outbreak Samples from Southern Thailand. Jpn. J. Infect. Dis. 2019, 72, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, S.; Khorami, N.; Ganji, Z.F.; Sepahian, N.; Malmasi, A.A.; Gouya, M.M.; Djadid, N.D. Leptospira wolffii, a potential new pathogenic Leptospira species detected in human, sheep and dog. Infect. Genet. Evol. 2010, 10, 273–277. [Google Scholar] [CrossRef]
- Balamurugan, V.; Gangadhar, N.L.; Mohandoss, N.; Thirumalesh, S.R.; Dhar, M.; Shome, R.; Krishnamoorthy, P.; Prabhudas, K.; Rahman, H. Characterization of leptospira isolates from animals and humans: Phylogenetic analysis identifies the prevalence of intermediate species in India. Springerplus 2013, 2, 362. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, S.; Sepahian, N.; Afsharpad, M.; Esfandiari, B.; Ziapour, P.; Djadid, N.D. Molecular epidemiology of leptospirosis in northern Iran by nested polymerase chain reaction/restriction fragment length polymorphism and sequencing methods. Am. J. Trop. Med. Hyg. 2010, 82, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Slack, A.T.; Kalambaheti, T.; Symonds, M.L.; Dohnt, M.F.; Galloway, R.L.; Steigerwalt, A.G.; Chaicumpa, W.; Bunyaraksyotin, G.; Craig, S.; Harrower, B.J.; et al. Leptospira wolffii sp. nov., isolated from a human with suspected leptospirosis in Thailand. Int. J. Syst. Evol. Microbiol. 2008, 58, 2305–2308. [Google Scholar] [CrossRef]
- Chiriboga, J.; Barragan, V.; Arroyo, G.; Sosa, A.; Birdsell, D.N.; España, K.; Mora, A.; Espín, E.; Mejía, M.E.; Morales, M.; et al. High prevalence of intermediate Leptospira spp. DNA in febrile humans from urban and rural Ecuador. Emerg. Infect. Dis. 2015, 21, 2141. [Google Scholar] [CrossRef]
- Ganoza, C.A.; Matthias, M.A.; Collins-Richards, D.; Brouwer, K.C.; Cunningham, C.B.; Segura, E.R.; Gilman, R.H.; Gotuzzo, E.; Vinetz, J.M. Determining risk for severe leptospirosis by molecular analysis of environmental surface waters for pathogenic Leptospira. PLoS Med. 2006, 3, e308. [Google Scholar] [CrossRef]
- Matthias, M.A.; Ricaldi, J.N.; Cespedes, M.; Diaz, M.M.; Galloway, R.L.; Saito, M.; Steigerwalt, A.G.; Patra, K.P.; Ore, C.V.; Gotuzzo, E.; et al. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl. Trop. Dis. 2008, 2, e213. [Google Scholar] [CrossRef]
- Tsuboi, M.; Koizumi, N.; Hayakawa, K.; Kanagawa, S.; Ohmagari, N.; Kato, Y. Imported Leptospira licerasiae infection in traveler returning to Japan from Brazil. Emerg. Infect. Dis. 2017, 23, 548. [Google Scholar] [CrossRef]
- Suwannarong, K.; Chapman, R.S. Characteristics associated with contact with rodents in, around, and outside homes in Khon Kaen province, Thailand. Am. J. Trop. Med. Hyg. 2015, 92, 784–790. [Google Scholar] [CrossRef]
- Suwancharoen, D.; Indrakamhang, P.; Neramitmansook, P.; Tangkanakul, W. Serological survey of Leptospiral antibodies in livestock in 5 northeastern provinces. J. Thai Vet. Med Assoc. 2000, 51, 9–18. [Google Scholar]
- Suwancharoen, D.; Chaisakdanugull, Y.; Thanapongtharm, W.; Yoshida, S. Serological survey of leptospirosis in livestock in Thailand. Epidemiol. Infect. 2013, 141, 2269–2277. [Google Scholar] [CrossRef] [PubMed]
- Wongpanit, K.; Suwanacharoen, D.; Srikram, A. Serological survey of leptospirosis in Thai swamp buffalo (Bubalus bubalis) in Sakon Nakhon province, Thailand. Agric. Nat. Resour. 2012, 46, 736–741. [Google Scholar]
- Suwancharoen, D.; Limlertvatee, S.; Chetiyawan, P.; Tongpan, P.; Sangkaew, N.; Sawaddee, Y.; Inthakan, K.; Wiratsudakul, A. A nationwide survey of pathogenic leptospires in urine of cattle and buffaloes by Loop-mediated isothermal amplification (LAMP) method in Thailand, 2011–2013. J. Vet. Med Sci. 2016, 15-0493. [Google Scholar] [CrossRef] [PubMed]
- Kurilung, A.; Chanchaithong, P.; Lugsomya, K.; Niyomtham, W.; Wuthiekanun, V.; Prapasarakul, N. Molecular detection and isolation of pathogenic Leptospira from asymptomatic humans, domestic animals and water sources in Nan province, a rural area of Thailand. Res. Vet. Sci. 2017, 115, 146–154. [Google Scholar] [CrossRef] [PubMed]
| Average Annual Incidence of Human Cases Per 100,000 | Number of Trapped Rodents (%) | Percentage of Trap Success | Leptospira Prevalence (%) | |||
|---|---|---|---|---|---|---|
| All | Linte 1 | Lborg 1 | ||||
| Provinces | ||||||
| Burirum | 23.54 | 320 (64.6) | 15.72 | 11 (3.4) | 8 (2.5) | 3 (0.9) |
| Surin | 29.63 | 43 (8.7) | 15.47 | 1 (2.3) | 1 (2.3) | 0 |
| Sisaket | 26.97 | 84 (17.0) | 32.85 | 6 (7.1) | 6 (7.1) | 0 |
| Kalasin | 19.28 | 48 (9.7) | 17.44 | 0 | 0 | 0 |
| Rodent Species | ||||||
| Rattus exulans | 246 (49.7) | 8.27 | 9 (3.6) | 6 (2.4) | 3 (1.2) | |
| Rattus rattus | 187 (37.8) | 6.22 | 6 (3.2) | 6 (3.2) | 0 | |
| Bandicota indica | 21 (4.2) | 0.68 | 3 (14.3) | 3 (14.3) | 0 | |
| Mus cervicolor | 14 (2.8) | 0.45 | 0 | 0 | 0 | |
| Mus caroli | 13 (2.6) | 0.42 | 0 | 0 | 0 | |
| Rattus losea | 5 (1.0) | 0.16 | 0 | 0 | 0 | |
| Rattus argentiventer | 4 (0.8) | 0.13 | 0 | 0 | 0 | |
| Bandicota savilei | 3 (0.6) | 0.1 | 0 | 0 | 0 | |
| Menetes berdmorei | 1 (0.2) | 0.03 | 0 | 0 | 0 | |
| Suncus murinus | 1 (0.2) | 0.03 | 0 | 0 | 0 | |
| Total | 495 (100) | 17.4 | 18 (3.6) | 15 (3.0) | 3 (0.6) | |
| Rodent Species | Provinces | |||
|---|---|---|---|---|
| Burirum | Surin | Sisaket | Kalasin | |
| Rattus exulans | 9/199 (4.5%) | 0/23 | 0/11 | 0/13 |
| Rattus rattus | 1/77 (1.3%) | 1/11 (9.0%) | 4/64 (6.3%) | 0/35 |
| Bandicota indica | 1/11 (9.0%) | 0/6 | 2/4 (50.0%) | 0/0 |
| Mus cervicolor | 0/14 | 0/0 | 0/0 | 0/0 |
| Mus caroli | 0/12 | 0/0 | 0/1 | 0/0 |
| Rattus losea | 0/4 | 0/0 | 0/1 | 0/0 |
| Rattus argentiventer | 0/1 | 0/0 | 0/3 | 0/0 |
| Bandicota savilei | 0/0 | 0/3 | 0/0 | 0/0 |
| Menetes berdmorei | 0/1 | 0/0 | 0/0 | 0/0 |
| Suncus murinus | 0/1 | 0/0 | 0/0 | 0/0 |
| Total | 11/320 (3.4%) | 1/43 (2.3%) | 6/84 (7.1%) | 0/48 (0%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krairojananan, P.; Thaipadungpanit, J.; Leepitakrat, S.; Monkanna, T.; Wanja, E.W.; Schuster, A.L.; Costa, F.; Poole-Smith, B.K.; McCardle, P.W. Low Prevalence of Leptospira Carriage in Rodents in Leptospirosis-Endemic Northeastern Thailand. Trop. Med. Infect. Dis. 2020, 5, 154. https://doi.org/10.3390/tropicalmed5040154
Krairojananan P, Thaipadungpanit J, Leepitakrat S, Monkanna T, Wanja EW, Schuster AL, Costa F, Poole-Smith BK, McCardle PW. Low Prevalence of Leptospira Carriage in Rodents in Leptospirosis-Endemic Northeastern Thailand. Tropical Medicine and Infectious Disease. 2020; 5(4):154. https://doi.org/10.3390/tropicalmed5040154
Chicago/Turabian StyleKrairojananan, Panadda, Janjira Thaipadungpanit, Surachai Leepitakrat, Taweesak Monkanna, Elizabeth W. Wanja, Anthony L. Schuster, Federico Costa, B. Katherine Poole-Smith, and Patrick W. McCardle. 2020. "Low Prevalence of Leptospira Carriage in Rodents in Leptospirosis-Endemic Northeastern Thailand" Tropical Medicine and Infectious Disease 5, no. 4: 154. https://doi.org/10.3390/tropicalmed5040154
APA StyleKrairojananan, P., Thaipadungpanit, J., Leepitakrat, S., Monkanna, T., Wanja, E. W., Schuster, A. L., Costa, F., Poole-Smith, B. K., & McCardle, P. W. (2020). Low Prevalence of Leptospira Carriage in Rodents in Leptospirosis-Endemic Northeastern Thailand. Tropical Medicine and Infectious Disease, 5(4), 154. https://doi.org/10.3390/tropicalmed5040154





