The Diagnostic Utility of PCR in FFPE Skin Biopsies with Inconclusive Histopathology for Suspected Cutaneous Leishmaniasis: A Pilot Study from Colombia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Histopathological Analysis
2.3. DNA Extraction from FFPE Tissues
2.4. PCR Amplification of the Miniexon Gene
2.5. PCR Amplification of rRNA Gene Internal Transcribed Spacer 1 (ITS1)
2.6. RFLP Analysis of the ITS1 PCR Amplicon
2.7. Ethical Considerations
2.8. Statistical Analysis
3. Results
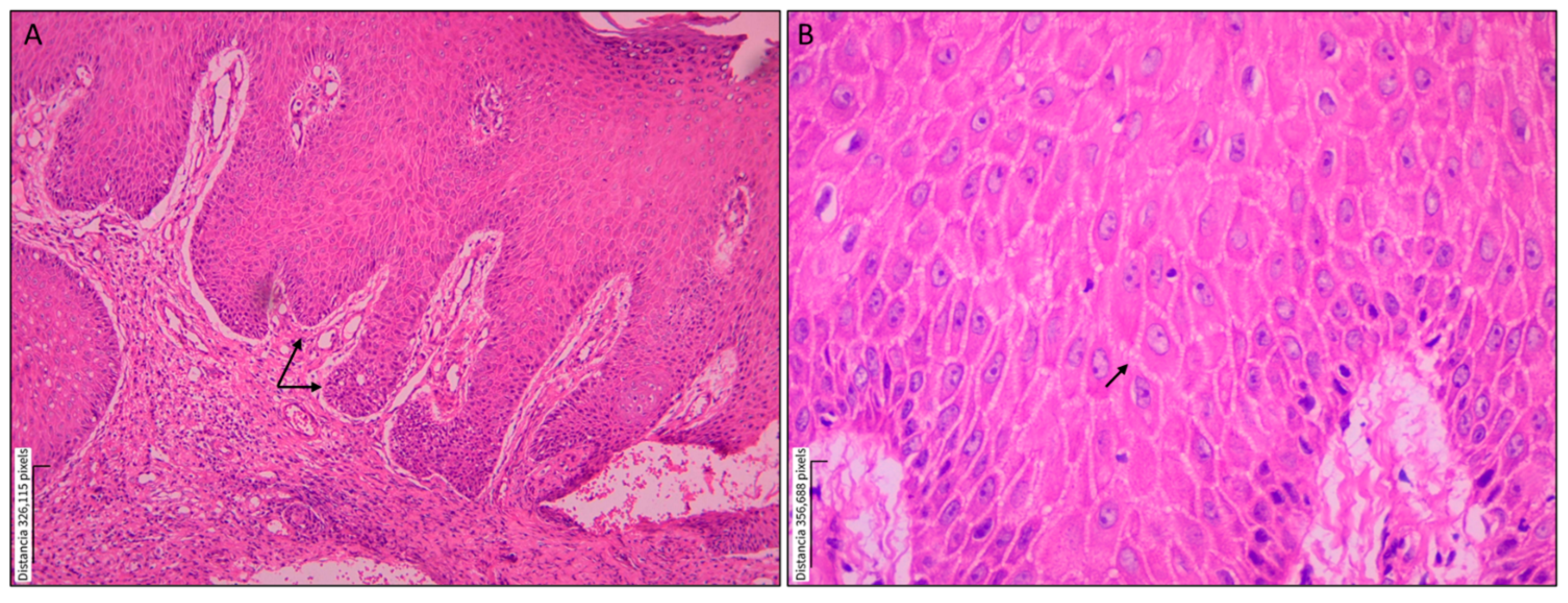
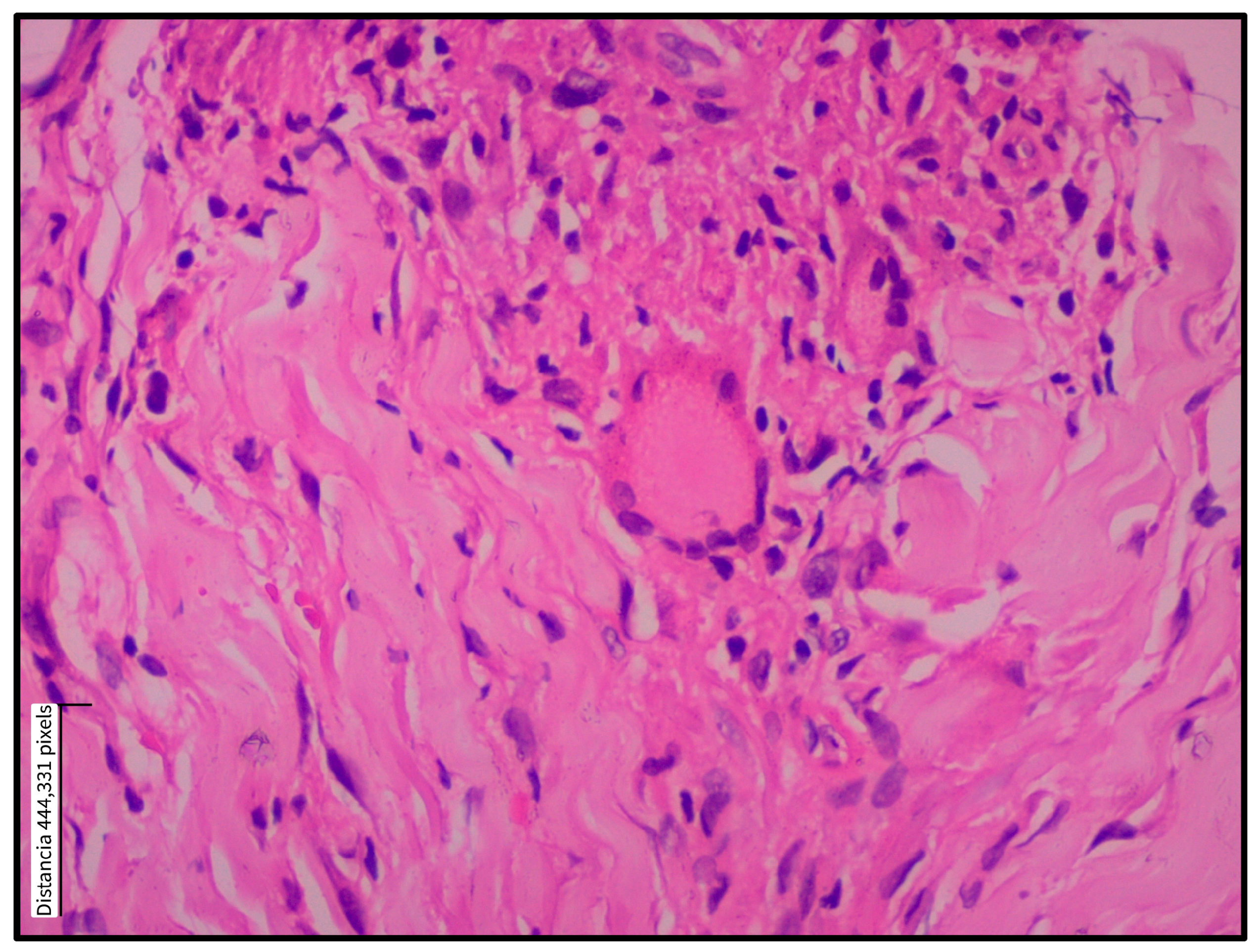
3.1. Histopathological Findings
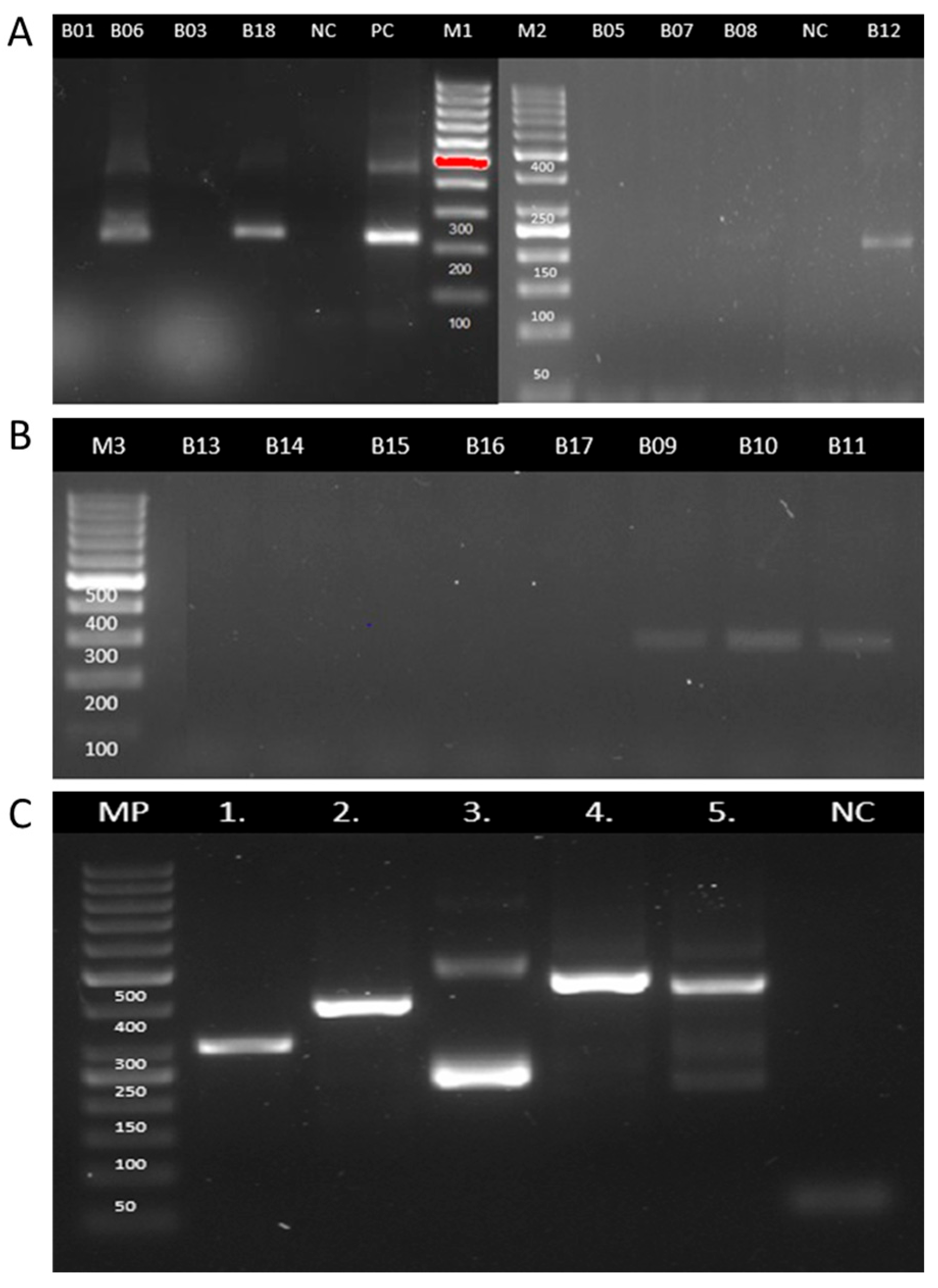
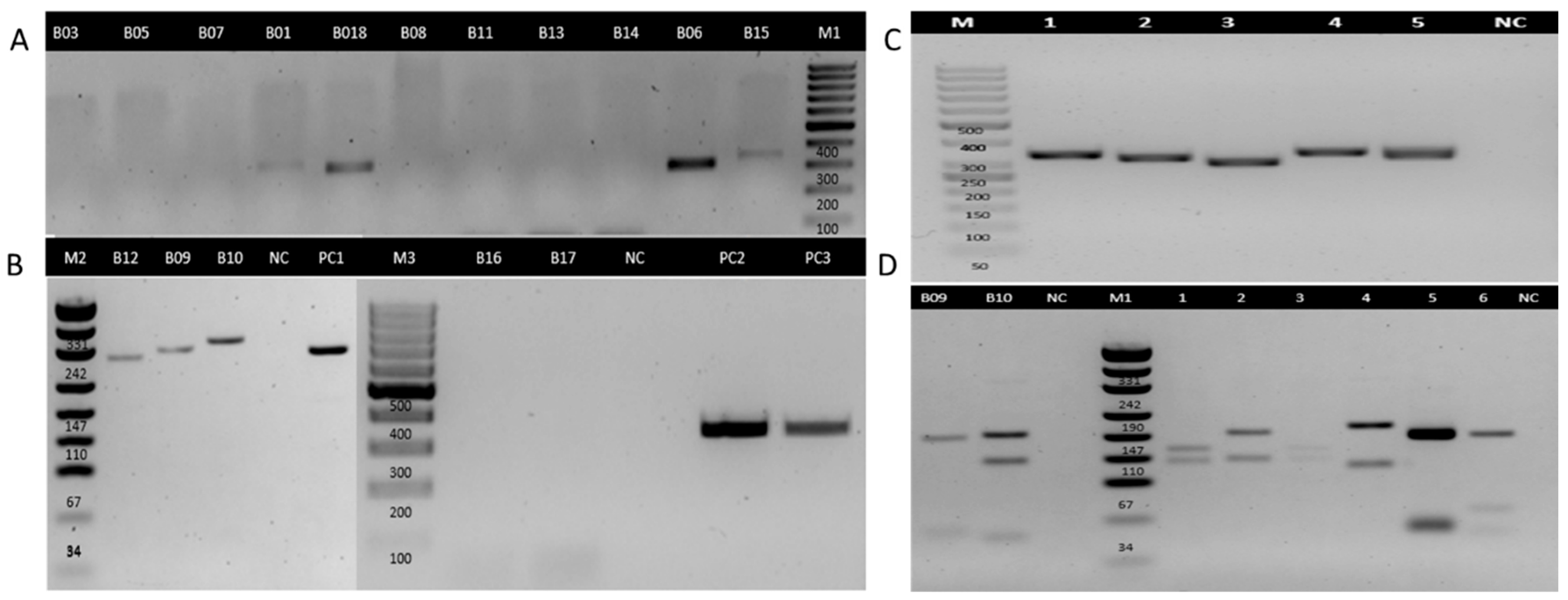
3.2. PCR Detection of Leishmania DNA
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arenas, R.; Torres-Guerrero, E.; Quintanilla-Cedillo, M.R.; Ruiz-Esmenjaud, J. Leishmaniasis: A review. F1000Research 2017, 6, 750. [Google Scholar]
- OMS. Leishmaniasis—PAHO/WHO|Pan American Health Organization. Pan American Health Organization. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 15 May 2025).
- Pan American Health Organization. Manual of Procedures for Surveillance and Control of Leishmaniasis in the Americas. World Health Organization. 2019. Available online: https://iris.paho.org/handle/10665.2/51838 (accessed on 15 May 2025).
- Bennis, I.; De Brouwere, V.; Belrhiti, Z.; Sahibi, H.; Boelaert, M. Psychosocial burden of localised cutaneous Leishmaniasis: A scoping review. BMC Public Health 2018, 18, 358. [Google Scholar] [CrossRef] [PubMed]
- Bilgic-Temel, A.; Murrell, D.F.; Uzun, S. Cutaneous leishmaniasis: A neglected disfiguring disease for women. Int. J. Women’s Dermatol. 2019, 5, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.S.; Lockwood, D.N.J. Cutaneous Leishmaniasis. Clin. Dermatol. 2007, 25, 203–211. Available online: https://www.sciencedirect.com/science/article/pii/S0738081X0600071X (accessed on 15 May 2025). [CrossRef] [PubMed]
- Komurcugil, I.; Nek, K.N. Cutaneous Leishmaniasis with Unusual Psoriasiform Presentation. J. Turk. Acad. Dermatol. 2022, 16, 53–55. [Google Scholar] [CrossRef]
- Guimarães, L.H.; Queiroz, A.; Silva, J.A.; Silva, S.C.; Magalhães, V.; Lago, E.L.; Machado, P.R.L.; Bacellar, O.; Wilson, M.E.; Beverley, S.M.; et al. Atypical Manifestations of Cutaneous Leishmaniasis in a Region Endemic for Leishmania braziliensis: Clinical, Immunological and Parasitological Aspects. PLoS Negl. Trop. Dis. 2016, 10, e0005100. [Google Scholar] [CrossRef]
- Meireles, C.B.; Maia, L.C.; Soares, G.C.; Teodoro, I.P.P.; Gadelha, M.D.S.V.; da Silva, C.G.L.; de Lima, M.A.P. Atypical presentations of cutaneous leishmaniasis: A systematic review. Acta Trop. 2017, 172, 240–254. [Google Scholar] [CrossRef]
- Piyasiri, S.B.; Dewasurendra, R.; Samaranayake, N.; Karunaweera, N. Diagnostic Tools for Cutaneous Leishmaniasis Caused by Leishmania donovani: A Narrative Review. Diagnostics 2023, 13, 2989. [Google Scholar] [CrossRef]
- Reina, A.M.; Mewa, J.C.; Calzada, J.E.; Saldaña, A. Characterization of Leishmania spp. Causing Cutaneous Lesions with a Negative Parasitological Diagnosis in Panama. Trop. Med. Infect. Dis. 2022, 7, 282. [Google Scholar] [CrossRef]
- Wijesinghe, H.D.; Wijesinghe, G.K.; Fernando, D.; de Silva, C. Immunopathology of Cutaneous Leishmaniasis in a Cohort of Sri Lankan Patients. Clin. Pathol. 2022, 15, 2632010X221134804. [Google Scholar] [CrossRef]
- de Vries, H.J.C.; Schallig, H.D. Cutaneous Leishmaniasis: A 2022 Updated Narrative Review into Diagnosis and Management Developments. Am. J. Clin. Dermatol. 2022, 23, 823–840. [Google Scholar] [CrossRef] [PubMed]
- Mesa, L.E.; Manrique, R.; Muskus, C.; Robledo, S.M. Test accuracy of polymerase chain reaction methods against conventional diagnostic techniques for cutaneous leishmaniasis (Cl) in patients with clinical or epidemiological suspicion of cl: Systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2020, 14, e0007981. [Google Scholar] [CrossRef]
- Greytak, S.R.; Engel, K.B.; Bass, B.P.; Moore, H.M. Accuracy of molecular data generated with FFPE Biospecimens: Lessons from the literature. Cancer Res. 2015, 75, 1541–1547. [Google Scholar] [CrossRef] [PubMed]
- Casaril, A.E.; de Oliveira, L.P.; Alonso, D.P.; de Oliveira, E.F.; Barrios, S.P.G.; Infran, J.d.O.M.; Fernandes, W.d.S.; Oshiro, E.T.; Ferreira, A.M.T.; Ribolla, P.E.M.; et al. Standardization of DNA extraction from sand flies: Application to genotyping by next generation sequencing. Exp. Parasitol. 2017, 177, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Marfurt, J.; Nasereddin, A.; Niederwieser, I.; Jaffe, C.L.; Beck, H.P.; Felger, I. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J. Clin. Microbiol. 2003, 41, 3147–3153. [Google Scholar] [CrossRef]
- El Tai, N.O.; El Fari, M.; Mauricio, I.; Miles, M.A.; Oskam, L.; El Safi, S.H.; Presber, W.H.; Schönian, G. Leishmania donovani: Intraspecific polymorphisms of Sudanese isolates revealed by PCR-based analyses and DNA sequencing. Exp. Parasitol. 2001, 97, 35–44. [Google Scholar] [CrossRef]
- Laskay, T.; Mikó, T.L.; Negesse, Y.; Solbach, W.; Röllinghoff, M.; Frommel, D. Detection of cutaneous leishmania infection in paraffin-embedded skin biopsies using the polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 1995, 89, 273–275. [Google Scholar] [CrossRef]
- Momeni, A.Z.; Yotsumoto, S.; Mehregan, D.R.; Mehregan, A.H.; Mehregan, D.A.; Aminjavaheri, M.; Fujiwara, H.; Tada, J. Chronic lupoid leishmaniasis: Evaluation by polymerase chain reaction. Arch. Dermatol. 1996, 132, 198–202. [Google Scholar] [CrossRef]
- Weigle, K.A.; Labrada, L.A.; Lozano, C.; Santrich, C.; Barker, D.C. PCR-based diagnosis of acute and chronic cutaneous leishmaniasis caused by Leishmania (Viannia). J. Clin. Microbiol. 2002, 40, 601–606. [Google Scholar] [CrossRef]
- Safaei, A.; Motazedian, M.H.; Vasei, M. Polymerase chain reaction for diagnosis of cutaneous leishmaniasis in histologically positive, suspicious and negative skin biopsies. Dermatology 2002, 205, 18–24. [Google Scholar] [CrossRef]
- Lanús, E.C.; Piñero, J.E.; González, A.C.; Valladares, B.; De Grosso, M.L.; Salomón, O.D. Detection of Leishmania braziliensis in human paraffin-embedded tissues from Tucumán, Argentina by polymerase chain reaction. Memórias Do Inst. Oswaldo Cruz 2005, 100, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Toz, S.O.; Culha, G.; Zeyrek, F.Y.; Ertabaklar, H.; Alkan, M.Z.; Vardarlı, A.T.; Gunduz, C.; Ozbel, Y.; Louzir, H. A Real-Time ITS1-PCR Based Method in the Diagnosis and Species Identification of Leishmania Parasite from Human and Dog Clinical Samples in Turkey. PLoS Negl. Trop. Dis. 2013, 7, e2205. [Google Scholar] [CrossRef]
- Dietrich, D.; Uhl, B.; Sailer, V.; Holmes, E.E.; Jung, M.; Meller, S.; Kristiansen, G.; Castresana, J.S. Improved PCR Performance Using Template DNA from Formalin-Fixed and Paraffin-Embedded Tissues by Overcoming PCR Inhibition. PLoS ONE 2013, 8, e77771. [Google Scholar] [CrossRef]
- Gebhardt, M.; Ertas, B.; Falk, T.M.; Blödorn-Schlicht, N.; Metze, D.; Böer-Auer, A. Fast, sensitive and specific diagnosis of infections with Leishmania spp. in formalin-fixed, paraffin-embedded skin biopsies by cytochrome b polymerase chain reaction. Br. J. Dermatol. 2015, 173, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Yuil, J.M.R.; Saldaña, A.; Calzada, J.; Arias, J.; Díaz, R.; González, K. Comparación entre histopatología y PCR, para diagnóstico de leishmaniasis tegumentaria. Dermatol. Cosmet. Medica Y Quir. 2012, 10, 13–20. [Google Scholar]
- van der Auwera, G.; Dujardina, J.C. Species typing in dermal leishmaniasis. Clin. Microbiol. Rev. 2015, 28, 265–294. [Google Scholar] [CrossRef]
- Chatterjee, D.; Bhattacharjee, R.; Saikia, U.N. Non-infectious granulomatous dermatoses: A pathologist’s perspective. Indian Dermatol. Online J. 2021, 12, 515–528. [Google Scholar] [CrossRef]
- Babouee Flury, B.; Weisser, M.; Prince, S.S.; Bubendorf, L.; Battegay, M.; Frei, R.; Goldenberger, D. Performances of two different panfungal PCRs to detect mould DNA in formalin-fixed paraffin-embedded tissue: What are the limiting factors? BMC Infect. Dis. 2014, 14, 692. [Google Scholar] [CrossRef]
- Braun-Falco, M.; Schempp, W.; Weyers, W. Molecular diagnosis in dermatopathology: What makes sense, and what doesn’t. Exp. Dermatol. 2008, 18, 12–23. [Google Scholar] [CrossRef]
- Thakur, S.; Joshi, J.; Kaur, S. Leishmaniasis diagnosis: An update on the use of parasitological, immunological and molecular methods. J. Parasit. Dis. 2020, 44, 253–272. [Google Scholar] [CrossRef]

| Characteristic | Category | n = 16 (%) |
|---|---|---|
| Sex | Male | 10 (62.5%) |
| Female | 6 (37.5%) | |
| Age group by sex | ||
| Male (n = 10) | Children (0–5 yrs) | 2 (20.0%) |
| Adolescents (10–17 yrs) | 1 (10.0%) | |
| Adults (18–59 yrs) | 4 (40.0%) | |
| Older adults (60+) | 3 (30.0%) | |
| Female (n = 6) | Children (0–5 yrs) | 1 (16.7%) |
| Adolescents (10–17 yrs) | 1 (16.7%) | |
| Adults (18–59 yrs) | 3 (50.0%) | |
| Older adults (60+) | 1 (16.7%) | |
| Site of lesion | Lower extremities | 9 (56.2%) |
| Upper extremities | 6 (37.5%) | |
| Head and face | 1 (6.3%) |
| Tissue Level | Histological Feature | n = 16 (%) | |
|---|---|---|---|
| Epidermis | Ulceration | 14 (87.5) | |
| Acanthosis | 15 (93.8) | ||
| Pseudoepitheliomatous hyperplasia | 14 (87.5) | ||
| Spongiosis | 13 (81.3) | ||
| Parakeratosis | 13 (81.3) | ||
| Transepidermal migration | 8 (50.0) | ||
| Dermis | Inflammation | 15 (93.8) | |
| Granulomatous pattern | 10 (62.5) | ||
| Fibrosis | 10 (62.5) | ||
| Vascular proliferation | 13 (81.3) | ||
| Vascular type | Telangiectasias | 6 (37.5) | |
| Prominent endothelium | 8 (50.0) | ||
| Inflammatory cell type | Abundant | Scant | |
| Plasma cells | 11 (68.8) | 5 (31.3) | |
| Lymphocytes | 13 (81.3) | 3 (18.8) | |
| Histiocytes | 6 (37.5) | 10 (62.5) | |
| Infiltrate classification | Mixed-type granuloma (100) | ||
| Infiltrate distribution | Perivascular | 0 (0) | |
| Interstitial | 0 (0) | ||
| Diffuse | 14 (87.5) | ||
| Nodular | 2 (12.5) | ||
| Granulomas | Tuberculoid | 3 (18.8) | |
| Epithelioid | 9 (56.3) | ||
| Sarcoid | 0 (0) | ||
| Giant cells | Multinucleated | 3 (18.8) | |
| Langhans-type multinucleated | 6 (37.5) | ||
| Histopathological Findings | Molecular Testing | ||||||
|---|---|---|---|---|---|---|---|
| Patient | Date of Sample Collection | Case Definition | Miniexon | ITS1 | Molecular Characterization | ||
| PCR | Subgenus * | PCR | RFLP ** | ||||
| B01 | 2024 | Suggestive | Negative | NA | Positive | NA | Leishmania spp. |
| B03 | 2021 | Suggestive | Negative | NA | Negative | NA | NA |
| B05 | 2021 | Suggestive | Negative | NA | Negative | NA | NA |
| B06 | 2023 | Suggestive | Positive | L. Viannia | Positive | NA | L. Viannia |
| B07 | 2021 | Suggestive | Negative | NA | Negative | NA | NA |
| B08 | 2021 | Suggestive | Negative | NA | Negative | NA | NA |
| B09 | 2021 | Suggestive | Positive | L. Leishmania | Positive | L. Leishmania | L. Leishmania |
| B10 | 2023 | Suggestive | Positive | L. Leishmania | Positive | L. Leishmania | L. Leishmania |
| B11 | 2023 | Suggestive | Positive | L. Leishmania | Negative | NA | L. Leishmania |
| B12 | 2024 | Suggestive | Positive | L. Viannia | Positive | NA | L. Viannia |
| B13 | 2021 | Suggestive | Negative | NA | Negative | NA | NA |
| B14 | 2021 | Suggestive | Negative | NA | Negative | NA | NA |
| B15 | 2024 | Suggestive | Negative | NA | Positive | NA | Leishmania spp. |
| B16 | 2024 | Suggestive | Negative | NA | Negative | NA | NA |
| B17 | 2024 | Suggestive | Negative | NA | Negative | NA | NA |
| B18 | 2024 | Suggestive | Positive | L. Viannia | Positive | NA | L. Viannia |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mantilla, J.C.; Bueno, N.A.; Alvarez, J.P.; López, M.P.; Díaz, M.L. The Diagnostic Utility of PCR in FFPE Skin Biopsies with Inconclusive Histopathology for Suspected Cutaneous Leishmaniasis: A Pilot Study from Colombia. Trop. Med. Infect. Dis. 2025, 10, 232. https://doi.org/10.3390/tropicalmed10080232
Mantilla JC, Bueno NA, Alvarez JP, López MP, Díaz ML. The Diagnostic Utility of PCR in FFPE Skin Biopsies with Inconclusive Histopathology for Suspected Cutaneous Leishmaniasis: A Pilot Study from Colombia. Tropical Medicine and Infectious Disease. 2025; 10(8):232. https://doi.org/10.3390/tropicalmed10080232
Chicago/Turabian StyleMantilla, Julio César, Nathalia Andrea Bueno, Juan Pablo Alvarez, Maria Paula López, and Martha Lucía Díaz. 2025. "The Diagnostic Utility of PCR in FFPE Skin Biopsies with Inconclusive Histopathology for Suspected Cutaneous Leishmaniasis: A Pilot Study from Colombia" Tropical Medicine and Infectious Disease 10, no. 8: 232. https://doi.org/10.3390/tropicalmed10080232
APA StyleMantilla, J. C., Bueno, N. A., Alvarez, J. P., López, M. P., & Díaz, M. L. (2025). The Diagnostic Utility of PCR in FFPE Skin Biopsies with Inconclusive Histopathology for Suspected Cutaneous Leishmaniasis: A Pilot Study from Colombia. Tropical Medicine and Infectious Disease, 10(8), 232. https://doi.org/10.3390/tropicalmed10080232







