Deltamethrin Selection Drives Transcriptomic Changes in Detoxification, Immune, and Cuticle Genes in Aedes aegypti
Abstract
1. Introduction
2. Materials and Methods
2.1. Aedes aegypti Populations
2.2. Bioassays and Deltamethrin Selection
2.3. RNA and Library Preparation
2.4. Differential Gene Expression Analysis
2.5. Gene Ontology Annotation and Functional Enrichment Analysis
3. Results
3.1. Bioassays and Selection with Deltamethrin
3.2. Sequencing, Alignment, and Read Quantification
3.3. Data Exploration
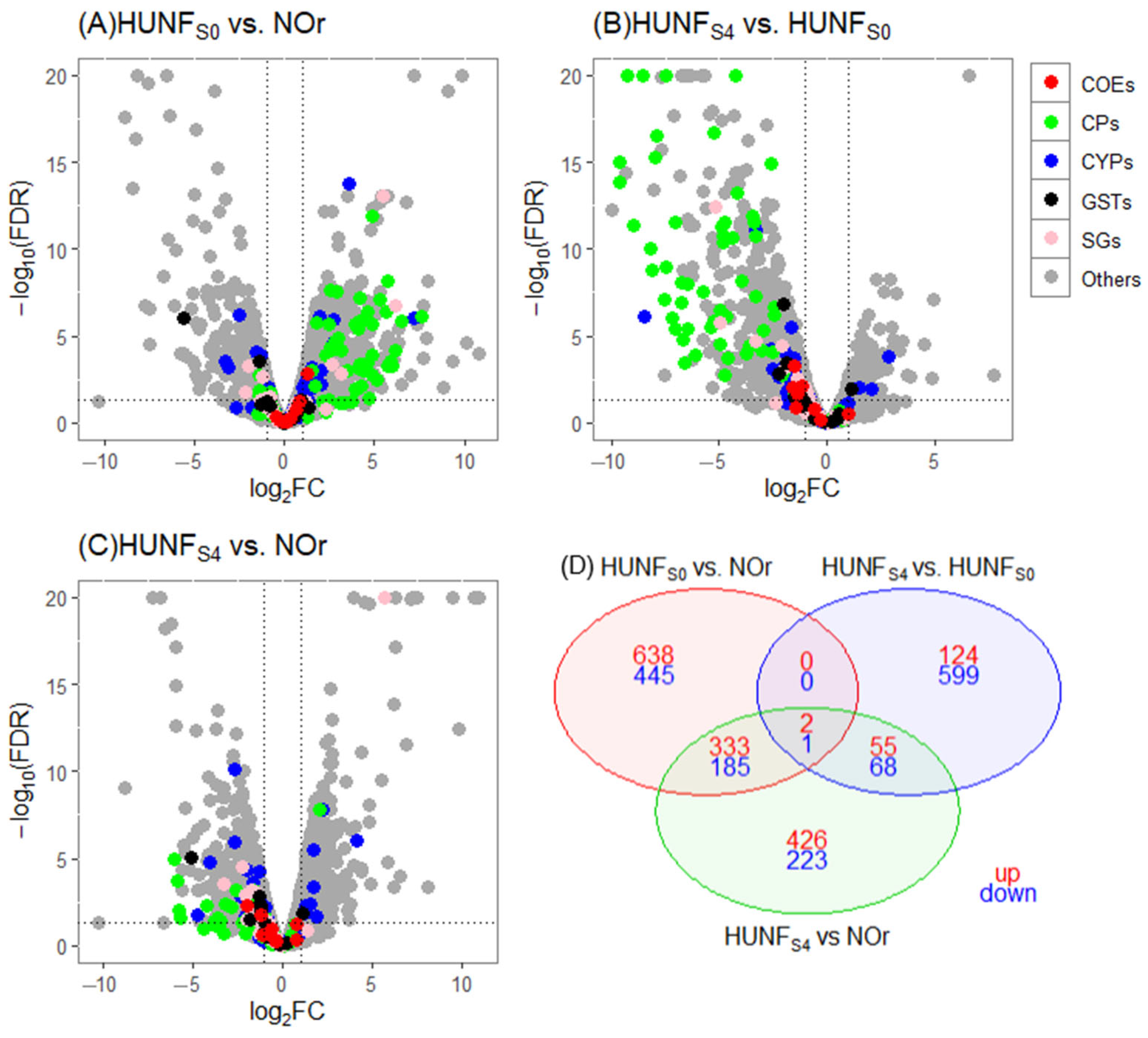
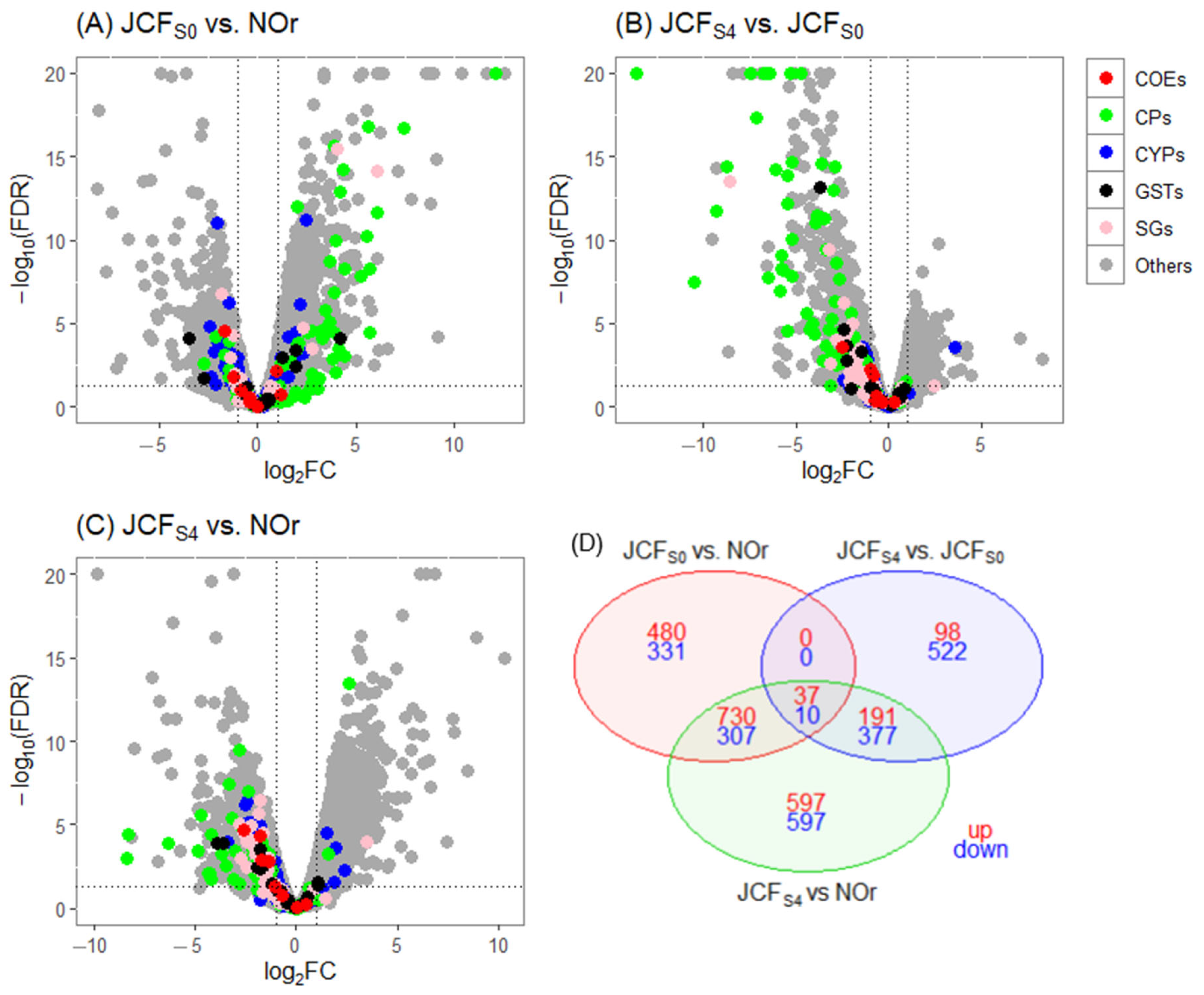
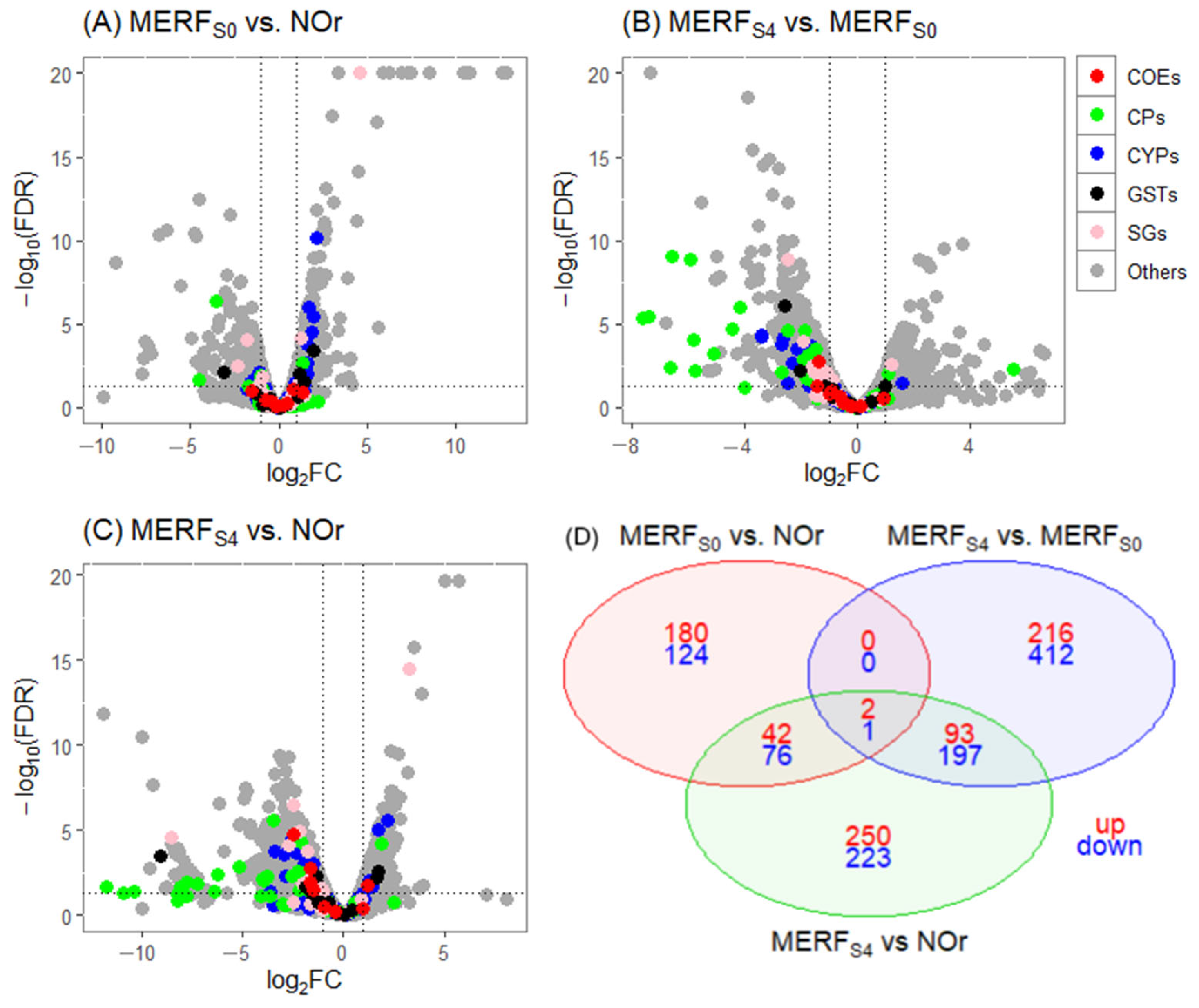
3.4. Differentially Expressed Genes
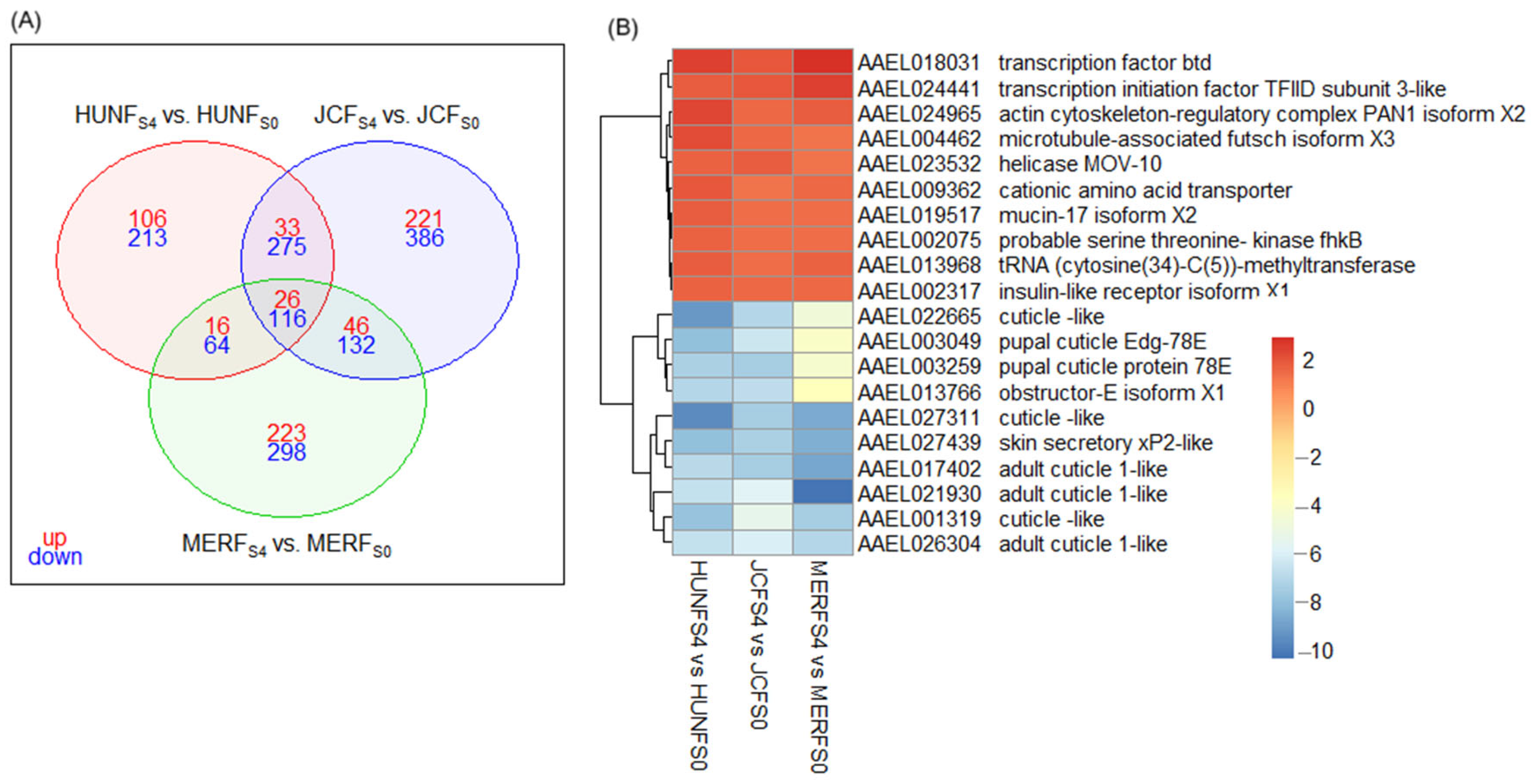
3.5. Gene Ontology Annotation and Enrichment Analysis
4. Discussion
4.1. Baseline Resistance Profiles and Gene Expression in FS0 vs. NOr
4.2. Transcriptomic Responses to Deltamethrin Selection in FS4 vs. FS0
4.3. Shared Patterns and Functional Responses in FS4
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- World Health Organization (WHO). Global Vector Control Response 2017–2030; World Health Organization: Geneva, Switzerland, 2017; Available online: https://www.who.int/publications/i/item/9789241512978 (accessed on 8 January 2025).
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.B.; Kasai, S.; Scott, J.G. Pyrethroid resistance in Aedes aegypti and Aedes albopictus: Important mosquito vectors of human diseases. Pestic. Biochem. Physiol. 2016, 133, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Huber, J.H.; Childs, M.L.; Caldwell, J.M.; Mordecai, E.A. Seasonal temperature variation influences climate suitability for dengue, chikungunya, and Zika transmission. PLoS Negl. Trop. Dis. 2018, 12, e0006451. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Matano, L.; Monroy-Muñoz, I.E.; Pardavé-Alejandre, H.D.; Uribe-Noguez, L.A.; Hernández-Cueto, M.L.A.; Rojas-Mendoza, T.; Santacruz-Tinoco, C.E.; Grajales-Muñiz, C.; Muñoz-Medina, J.E. Impact of the introduction of chikungunya and Zika viruses on the incidence of dengue in endemic zones of Mexico. PLoS Negl. Trop. Dis. 2021, 15, e0009922. [Google Scholar] [CrossRef]
- Pan American Health Organization. Technical Document for the Implementation of Interventions Based on Generic Operational Scenarios for Aedes aegypti Control; Pan American Health Organization: Washington, DC, USA, 2019; Available online: https://iris.paho.org/handle/10665.2/51652 (accessed on 12 January 2025).
- World Health Organization (WHO). Guidelines for Procuring Public Health Pesticides; World Health Organization: Geneva, Switzerland, 2012; Available online: https://www.who.int/publications/i/item/9789241503426 (accessed on 16 November 2024).
- Moyes, C.L.; Vontas, J.; Martins, A.J.; Ng, L.C.; Koou, S.Y.; Dusfour, I.; Raghavendra, K.; Pinto, J.; Corbel, V.; David, J.P.; et al. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl. Trop. Dis. 2017, 11, e0005625. [Google Scholar] [CrossRef]
- Ray, D.E.; Fry, J.R. A reassessment of the neurotoxicity of pyrethroid insecticides. Pharmacol. Ther. 2006, 111, 174–193. [Google Scholar] [CrossRef]
- Wolansky, M.J.; Harrill, J.A. Neurobehavioral toxicology of pyrethroid insecticides in adult animals: A critical review. Neurotoxicol. Teratol. 2008, 30, 55–78. [Google Scholar] [CrossRef]
- Flores, A.E.; Ponce, G.; Silva, B.G.; Gutierrez, S.M.; Bobadilla, C.; Lopez, B.; Mercado, R.; Black WC, I.V. Widespread cross-resistance to pyrethroids in Aedes aegypti (Diptera: Culicidae) from Veracruz State, Mexico. J. Econ. Entomol. 2013, 106, 959–969. [Google Scholar] [CrossRef]
- Aponte, H.A.; Penilla, R.P.; Dzul-Manzanilla, F.; Che-Mendoza, A.; López, A.D.; Solis, F.; Manrique-Saide, P.; Ranson, H.; Lenhart, A.; McCall, P.J.; et al. The pyrethroid resistance status and mechanisms in Aedes aegypti from the Guerrero State, Mexico. Pestic. Biochem. Physiol. 2013, 107, 226–234. [Google Scholar] [CrossRef]
- Lopez-Monroy, B.; Gutierrez-Rodriguez, S.M.; Villanueva-Segura, O.K.; Ponce-Garcia, G.; Morales-Forcada, F.; Alvarez, L.C.; Flores, A.E. Frequency and intensity of pyrethroid resistance through the CDC bottle bioassay and their association with the frequency of kdr mutations in Aedes aegypti (Diptera: Culicidae) from Mexico. Pest Manag. Sci. 2018, 74, 2176–2184. [Google Scholar] [CrossRef]
- López-Solís, A.D.; Castillo-Vera, A.; Cisneros, J.; Solís-Santoyo, F.; Penilla-Navarro, R.P.; Black WC, I.V.; Torres-Estrada, J.L.; Rodríguez, A.D. Resistencia a insecticidas en Aedes aegypti y Aedes albopictus (Diptera: Culicidae) de Tapachula, Chiapas, México. Salud Publica Mex. 2020, 62, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Perera, Y.; Ponce-Garcia, G.; Villanueva-Segura, K.; Lopez-Monroy, B.; Rodríguez-Sanchez, I.P.; Lenhart, A.; Manrique-Saide, P.; Flores, A.E. Impact of deltamethrin selection on kdr mutations and insecticide detoxifying enzymes in Aedes aegypti from Mexico. Parasites Vectors 2020, 13, 224. [Google Scholar] [CrossRef] [PubMed]
- Solis-Santoyo, F.; Rodriguez, A.D.; Penilla-Navarro, R.P.; Sanchez, D.; Castillo-Vera, A.; Lopez-Solis, A.D.; Vazquez-Lopez, E.D.; Lozano, S.; Black WC, I.V.; Saavedra-Rodriguez, K. Insecticide resistance in Aedes aegypti from Tapachula, Mexico: Spatial variation and response to historical insecticide use. PLoS Negl. Trop. Dis. 2021, 15, e0009746. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Du, Y.; Rinkevich, F.; Nomura, Y.; Xu, P.; Wang, L.; Silver, K.; Zhorov, B.S. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 2014, 50, 1–17. [Google Scholar] [CrossRef]
- Du, Y.; Nomura, Y.; Satar, G.; Hu, Z.; Nauen, R.; He, S.Y.; Zhorov, B.S.; Dong, K. Molecular evidence for dual pyrethroid-receptor sites on a mosquito sodium channel. Proc. Natl. Acad. Sci. USA 2013, 110, 11785–11790. [Google Scholar] [CrossRef]
- Haddi, K.; Tomé, H.V.V.; Du, Y.; Valbon, W.R.; Nomura, Y.; Martins, G.F.; Dong, K.; Oliveira, E.E. Detection of a new pyrethroid resistance mutation (V410L) in the sodium channel of Aedes aegypti: A potential challenge for mosquito control. Sci. Rep. 2017, 7, 46549. [Google Scholar] [CrossRef]
- Saavedra-Rodriguez, K.; Maloof, F.V.; Campbell, C.L.; Garcia-Rejon, J.; Lenhart, A.; Penilla, P.; Rodriguez, A.; Sandoval, A.A.; Flores, A.E.; Ponce, G.; et al. Parallel evolution of vgsc mutations at domains IS6, IIS6, and IIIS6 in pyrethroid resistant Aedes aegypti from Mexico. Sci. Rep. 2018, 8, 6747. [Google Scholar] [CrossRef]
- Liu, N. Insecticide resistance in mosquitoes: Impact, mechanisms, and research directions. Annu. Rev. Entomol. 2015, 60, 537–559. [Google Scholar] [CrossRef]
- Faucon, F.; Gaude, T.; Dusfour, I.; Navratil, V.; Corbel, V.; Juntarajumnong, W.; Girod, R.; Poupardin, R.; Boyer, F.; Reynaud, S.; et al. In the hunt for genomic markers of metabolic resistance to pyrethroids in the mosquito Aedes aegypti: An integrated next-generation sequencing approach. PLoS Negl. Trop. Dis. 2017, 11, e0005526. [Google Scholar] [CrossRef]
- Flores, A.E.; Albeldaño-Vázquez, W.; Fernandez Salas, I.; Badii, M.H.; Loaiza Becerra, H.; Ponce Garcia, G.; Lozano Fuentes, S.; Brogdon, W.G.; Black WC, I.V.; Beaty, B. Elevated α-esterase levels associated with permethrin tolerance in Aedes aegypti (L.) from Baja California, Mexico. Pestic. Biochem. Physiol. 2005, 82, 66–78. [Google Scholar] [CrossRef]
- Flores, A.E.; Salomon Grajales, J.; Fernandez Salas, I.; Garcia, G.P.; Becerra, M.H.; Lozano, S.; Brogdon, W.G.; Black WC, I.V.; Beaty, B. Mechanisms of insecticide resistance in field populations of Aedes aegypti (L.) from Quintana Roo, southern Mexico. J. Am. Mosq. Control Assoc. 2006, 22, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.E.; Reyes Solis, G.; Fernandez Salas, I.; Sanchez Ramos, F.J.; Ponce Garcia, G. Resistance to permethrin in Aedes aegypti (L.) in northern Mexico. Southwest. Entomol. 2009, 34, 167–177. [Google Scholar] [CrossRef]
- Saavedra-Rodriguez, K.; Strode, C.; Flores-Suarez, A.; Fernandez-Salas, I.; Ranson, H.; Hemingway, J.; Black WC, I.V. QTL mapping of gene regions controlling permethrin resistance in the mosquito Aedes aegypti. Genetics 2008, 180, 1137–1152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lopez, B.; Ponce, G.; Gonzalez, J.A.; Gutierrez, S.M.; Villanueva, O.K.; Gonzalez, G.; Bobadilla, C.; Rodriguez, I.P.; Black WC, I.V.; Flores, A.E. Susceptibility to chlorpyrifos in pyrethroid-resistant populations of Aedes aegypti (Diptera: Culicidae) from Mexico. J. Med. Entomol. 2014, 51, 644–649. [Google Scholar] [CrossRef]
- Nkya, T.E.; Akhouayri, I.; Kisinza, W.; David, J.P. Impact of environment on mosquito response to pyrethroid insecticides: Facts, evidences and prospects. Insect Biochem. Mol. Biol. 2013, 43, 407–416. [Google Scholar] [CrossRef]
- Liao, C.; Upadhyay, A.; Liang, J.; Han, Q.; Li, J. 4-Dihydroxyphenylacetaldehyde synthase and cuticle formation in insects. Dev. Comp. Immunol. 2018, 83, 44–50. [Google Scholar] [CrossRef]
- Ren, Y.; Li, Y.; Ju, Y.; Zhang, W.; Wang, Y. Insect cuticle and insecticide development. Arch. Insect Biochem. Physiol. 2023, 114, e22057. [Google Scholar] [CrossRef]
- Wood, O.; Hanrahan, S.; Coetzee, M.; Koekemoer, L.; Brooke, B. Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus. Parasites Vectors 2010, 3, 67. [Google Scholar] [CrossRef]
- Ingham, V.A.; Anthousi, A.; Douris, V.; Harding, N.J.; Lycett, G.; Morris, M.; Vontas, J.; Ranson, H. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 2020, 577, 376–380. [Google Scholar] [CrossRef]
- Vieira, F.G.; Rozas, J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Hawkes, N.J.; McCarroll, L.; Ranson, H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem. Mol. Biol. 2004, 34, 653–665. [Google Scholar] [CrossRef]
- Campbell, C.L.; Saavedra-Rodriguez, K.; Kubik, T.D.; Lenhart, A.; Lozano-Fuentes, S.; Black WC, I.V. Vgsc-interacting proteins are genetically associated with pyrethroid resistance in Aedes aegypti. PLoS ONE 2019, 14, e0211497. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Coissac, E.; Melodelima, C.; Poupardin, R.; Riaz, M.A.; Chandor-Proust, A.; Reynaud, S. Transcriptome response to pollutants and insecticides in the dengue vector Aedes aegypti using next-generation sequencing technology. BMC Genom. 2010, 11, 216. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie-Impoinvil, L.; Weedall, G.D.; Lol, J.C.; Pinto, J.; Vizcaino, L.; Dzuris, N.; Riveron, J.; Padilla, N.; Wondji, C.; Lenhart, A. Contrasting patterns of gene expression indicate differing pyrethroid resistance mechanisms across the range of the New World malaria vector Anopheles albimanus. PLoS ONE 2019, 14, e0210586. [Google Scholar] [CrossRef]
- Brogdon, W.G.; McAllister, J.C. Simplification of adult mosquito bioassays through use of time-mortality determinations in glass bottles. J. Am. Mosq. Control Assoc. 1998, 14, 159–164. [Google Scholar]
- Lozano, F.S.; Saavedra, R.K.; Black WC, I.V.; Eisen, L. QCAL: A software application for the calculation of dose–response curves in insecticide resistance bioassays. J. Am. Mosq. Control Assoc. 2012, 28, 59–91. [Google Scholar] [CrossRef]
- Derilus, D.; Impoinvil, L.M.; Muturi, E.J.; McAllister, J.; Kenney, J.; Massey, S.E.; Hemme, R.; Kothera, L.; Lenhart, A. Comparative transcriptomic analysis of insecticide-resistant Aedes aegypti from Puerto Rico reveals insecticide-specific patterns of gene expression. Genes 2023, 14, 1626. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data (Version 0.11.2); Babraham Bioinformatics: Cambridge, UK, 2010; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 July 2022).
- Chen, Y.; McCarthy, D.; Ritchie, M.; Robinson, M.; Smyth, G. edgeR: Differential Analysis of Sequence Read Count Data User’s Guide. 2018. Available online: https://chagall.weill.cornell.edu/RNASEQcourse/edgeRUsersGuide-2018.pdf (accessed on 10 July 2022).
- Liao, Y.; Smyth, G.K.; Shi, W. Accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Chang, W.; Wickham, M.H. Package ‘ggplot2’: Create Elegant Data Visualisations Using the Grammar of Graphics, Version 2, 1–189. 2016. Available online: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed on 10 July 2022).
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’, R Package, Version 1. 2018. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 4 April 2025).
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed]
- Bonizzoni, M.; Dunn, W.A.; Campbell, C.L.; Olson, K.E.; Marinotti, O.; James, A.A. Strain variation in the transcriptome of the dengue fever vector, Aedes aegypti. G3 2012, 2, 103–114. [Google Scholar] [CrossRef]
- Zhao, L.; Becnel, J.J.; Clark, G.G.; Linthicum, K.J.; Chen, J.; Jin, X. Identification and expression profile of multiple genes in response to magnesium exposure in Culex quinquefasciatus larvae. J. Med. Entomol. 2010, 47, 1053–1061. [Google Scholar] [CrossRef]
- David, J.P.; Faucon, F.; Chandor-Proust, A.; Poupardin, R.; Riaz, M.A.; Bonin, A.; Navratil, V.; Reynaud, S. Comparative analysis of response to selection with three insecticides in the dengue mosquito Aedes aegypti using mRNA sequencing. BMC Genomics 2014, 15, 174. [Google Scholar] [CrossRef]
- Strode, C.; Wondji, C.S.; David, J.P.; Hawkes, N.J.; Lumjuan, N.; Nelson, D.R.; Drane, D.R.; Karunaratne, S.H.P.; Hemingway, J.; Black WC, I.V.; et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2008, 38, 113–123. [Google Scholar] [CrossRef]
- Poupardin, R.; Riaz, M.A.; Vontas, J.; David, J.P.; Reynaud, S. Transcription profiling of eleven cytochrome P450s potentially involved in xenobiotic metabolism in the mosquito Aedes aegypti. Insect Mol. Biol. 2010, 19, 185–193. [Google Scholar] [CrossRef]
- Saavedra-Rodríguez, K.; Flores Suarez, A.; Fernández Salas, I.; Strode, C.; Ranson, H.; Hemingway, J.; Black WC, I.V. Transcription of detoxification genes after permethrin selection in the mosquito Aedes aegypti. Insect Mol. Biol. 2012, 21, 61–77. [Google Scholar] [CrossRef]
- Epelboin, Y.; Wang, L.; Giai Gianetto, Q.; Choumet, V.; Gaborit, P.; Issaly, J.; Guidez, A.; Douché, T.; Chaze, T.; Matondo, M.; et al. CYP450 core involvement in multiple resistance strains of Aedes aegypti from French Guiana highlighted by proteomics, molecular and biochemical studies. PLoS ONE 2021, 16, e0243992. [Google Scholar] [CrossRef]
- Kubik, T.D.; Snell, T.K.; Saavedra-Rodriguez, K.; Wilusz, J.; Anderson, J.R.; Lozano-Fuentes, S.; Black WC, I.V.; Campbell, C.L. Aedes aegypti miRNA-33 modulates permethrin induced toxicity by regulating vgsc transcripts. Sci. Rep. 2021, 11, 7301. [Google Scholar] [CrossRef] [PubMed]
- Saavedra-Rodriguez, K.; Campbell, C.L.; Lozano, S.; Penilla-Navarro, P.; Lopez-Solis, A.; Solis-Santoyo, F.; Rodriguez, A.D.; Perera, R.; Black WC, I.V. Permethrin resistance in Aedes aegypti: Genomic variants that confer knockdown resistance, recovery, and death. PLoS Genet. 2021, 17, e1009606. [Google Scholar] [CrossRef] [PubMed]
- Cenaprece. Lista de Productos Recomendados para el Combate de Insectos Vectores a Partir de 2019; Cenaprece: Mexico City, Mexico, 2019; Available online: https://www.gob.mx/salud/cenaprece/documentos/lista-de-insumos-recomendados-por-el-cenaprece (accessed on 12 December 2020).
- Zoh, M.G.; Bonneville, J.M.; Laporte, F.; Tutagata, J.; Sadia, C.G.; Fodjo, B.K.; Mouhamadou, C.S.; McBeath, J.; Schmitt, F.; Horstmann, S.; et al. Deltamethrin and transfluthrin select for distinct transcriptomic responses in the malaria vector Anopheles gambiae. Malar. J. 2023, 22, 256. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Field, L.; Vontas, J. An overview of insecticide resistance. Science 2002, 298, 96–97. [Google Scholar] [CrossRef]
- Ffrench-Constant, R.H.; Daborn, P.J.; Le Goff, G. The genetics and genomics of insecticide resistance. Trends Genet. 2004, 20, 163–170. [Google Scholar] [CrossRef]
- Saavedra-Rodriguez, K.; Beaty, M.; Lozano-Fuentes, S.; Denham, S.; Garcia-Rejon, J.; Reyes-Solis, G.; Machain-Williams, C.; Loroño-Pino, M.A.; Flores-Suarez, A.; Ponce-Garcia, G.; et al. Local evolution of pyrethroid resistance offsets gene flow among Aedes aegypti collections in Yucatan State, Mexico. Am. J. Trop. Med. Hyg. 2015, 92, 201–209. [Google Scholar] [CrossRef][Green Version]
- Jacobs, E.; Chrissian, C.; Rankin-Turner, S.; Wear, M.; Camacho, E.; Broderick, N.A.; McMeniman, C.J.; Stark, R.E.; Casadevall, A. Cuticular profiling of insecticide resistant Aedes aegypti. Sci. Rep. 2023, 13, 10154. [Google Scholar] [CrossRef]
- Mejía, A.; Mejía-Jaramillo, A.M.; Fernandez, G.J.; Granada, Y.; Lowenberger, C.; Triana-Chávez, O. Long-term exposure to lambda-cyhalothrin reveals novel genes potentially involved in Aedes aegypti insecticide resistance. Insects 2025, 16, 106. [Google Scholar] [CrossRef]
- Bariami, V.; Jones, C.M.; Poupardin, R.; Vontas, J.; Ranson, H. Gene amplification, ABC transporters and cytochrome P450s: Unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti. PLoS Negl. Trop. Dis. 2012, 6, e1692. [Google Scholar] [CrossRef]
- Cattel, J.; Faucon, F.; Le Péron, B.; Sherpa, S.; Monchal, M.; Grillet, L.; Gaude, T.; Laporte, F.; Dusfour, I.; Reynaud, S.; et al. Combining genetic crosses and pool targeted DNA-seq for untangling genomic variations associated with resistance to multiple insecticides in the mosquito Aedes aegypti. Evol. Appl. 2020, 13, 303–317. [Google Scholar] [CrossRef]
- Namiki, T.; Niwa, R.; Sakudoh, T.; Shirai, K.I.; Takeuchi, H.; Kataoka, H. Cytochrome P450 CYP307A1/Spook: A regulator for ecdysone synthesis in insects. Biochem. Biophys. Res. Commun. 2005, 337, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Wang, Y.; Huang, Q.; Xu, Z.; Zhang, Y.; Hasnain, A.; Zhan, X.; He, Y.; Zhang, T.; Shen, L.; et al. Maf regulates the overexpression of CYP307A1, which is involved in the fitness advantage of bistrifluron-resistant Spodoptera litura (Fab.) (Noctuidae: Lepidoptera). Ecotoxicol. Environ. Saf. 2022, 234, 113425. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.; Rewitz, K.F.; Shinoda, T.; Itoyama, K.; Petryk, A.; Rybczynski, R.; Jarcho, M.; Warren, J.T.; Marqués, G.; Shimell, M.J.; et al. Spook and spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev. Biol. 2006, 298, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Mueller, C.R.; Fuchs, J.F.; Wessely, V.; Lan, Q.; Christensen, B.M. Regulatory mechanisms of chitin biosynthesis and roles of chitin in peritrophic matrix formation in the midgut of adult Aedes aegypti. Insect Biochem. Mol. Biol. 2006, 36, 1–9. [Google Scholar] [CrossRef]
- Shao, L.; Devenport, M.; Fujioka, H.; Ghosh, A.; Jacobs-Lorena, M. Identification and characterization of a novel peritrophic matrix protein, Ae-Aper50, and the microvillar membrane protein, AEG12, from the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2005, 35, 947–959. [Google Scholar] [CrossRef]
- Whiten, S.R.; Ray, W.K.; Helm, R.F.; Adelman, Z.N.; Weiss, B.L. Characterization of the adult Aedes aegypti early midgut peritrophic matrix proteome using LC-MS. PLoS ONE 2018, 13, e0194734. [Google Scholar] [CrossRef]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef]
- Rund, S.S.; Hou, T.Y.; Ward, S.M.; Collins, F.H.; Duffield, G.E. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2011, 108, E421–E430. [Google Scholar] [CrossRef]
- Chamnanya, S.; Kiddela, B.; Saingamsook, J.; Nachaiwieng, W.; Lumjuan, N.; Somboon, P.; Yanola, J. Overexpression of multiple cytochrome P450 genes with and without knockdown resistance mutations confers high resistance to deltamethrin in Culex quinquefasciatus. Infect. Dis. Poverty 2025, 14, 2. [Google Scholar] [CrossRef]
- Gonzalez-Santillan, F.J.; Contreras-Perera, Y.; Davila-Barboza, J.A.; Juache-Villagrana, A.E.; Gutierrez-Rodriguez, S.M.; Ponce-Garcia, G.; Lopez-Monroy, B.; Rodriguez-Sanchez, I.P.; Lenhart, A.E.; Mackenzie-Impoinvil, L.; et al. Fitness Cost of sequential selection with deltamethrin in Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 2022, 59, 930–939. [Google Scholar] [CrossRef]
- Tellam, R. The peritrophic matrix. In Biology of the Insect Midgut; Lehane, M.J., Billingsley, P.F., Eds.; Springer: Dordrecht, The Netherlands, 1996; pp. 86–114. [Google Scholar] [CrossRef]
- Morlais, S.; Severson, D.W. Identification of a polymorphic mucin-like gene expressed in the midgut of the mosquito, Aedes aegypti, using an integrated bulked segregant and differential display analysis. Genetics 2001, 158, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Klowden, M.J. Physiological Systems in Insects, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Bavithra, C.M.L.; Murugan, M.; Pavithran, S.; Naveena, K. Enthralling genetic regulatory mechanisms meddling insecticide resistance development in insects: Role of transcriptional and post-transcriptional events. Front. Mol. Biosci. 2023, 10, 1257859. [Google Scholar] [CrossRef]
- Mogilicherla, K.; Roy, A. Epigenetic regulations as drivers of insecticide resistance and resilience to climate change in arthropod pests. Front. Genet. 2023, 13, 1044980. [Google Scholar] [CrossRef]
- Xiao, H.; Ma, C.; Peng, R.; Xie, M. Insights into the role of non-coding RNAs in the development of insecticide resistance in insects. Front. Genet. 2024, 15, 1429411. [Google Scholar] [CrossRef] [PubMed]
- Spadar, A.; Collins, E.; Messenger, L.A.; Clark, T.G.; Campino, S. Uncovering the Genetic Diversity in Aedes aegypti Insecticide Resistance Genes through Global Comparative Genomics. Sci. Rep. 2024, 14, 13447. [Google Scholar] [CrossRef]
- Lisi, F.; Amichot, M.; Desneux, N.; Gatti, J.L.; Guedes, R.N.C.; Nazzi, F.; Pennacchio, F.; Russo, A.; Sánchez-Bayo, F.; Wang, X.; et al. Pesticide immunotoxicity on insects—Are agroecosystems at risk? Sci. Total Environ. 2024, 951, 175467. [Google Scholar] [CrossRef]
- Costa, C.; Briguglio, G.; Catanoso, R.; Giambò, F.; Polito, I.; Teodoro, M.; Fenga, C. New perspectives on cytokine pathways modulation by pesticide exposure. Curr. Opin. Toxicol. 2020, 19, 34–41. [Google Scholar] [CrossRef]
- Lee, G.H.; Choi, K.C. Adverse effects of pesticides on the functions of immune system. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2020, 235, 108789. [Google Scholar] [CrossRef]

| Strain | Generation | N 2 | LC50 (mg/Bottle) | CI 95% 3 | Slope ± SE 4 | RR50 6 |
|---|---|---|---|---|---|---|
| Merida | FS0 | 466 | 5.35 (4.73–6.56) | 4.73–6.56 | 1.06 (0.09) | 134 |
| FS4 | 501 | 7.73 (6.93–8.61) | 6.93–8.61 | 1.92 (0.17) | 193 | |
| Hunucma | FS0 | 424 | 0.25 (0.20–0.29) | 0.20–0.29 | 1.24 (0.12) | 6 |
| FS4 | 260 | 1.33 (1.16–1.52) | 1.16–1.52 | 2.45 (0.26) | 33 | |
| Jose Cardel | FS0 | 481 | 1.64 (1.46–1.83) | 1.46–1.83 | 1.89 (0.18) | 41 |
| FS4 | 420 | 2.26 (2.07–2.45) | 2.07–2.45 | 2.81 (0.25) | 57 | |
| New Orleans 5 | - | 492 | 0.04 (0.03–0.05) | 0.03–0.05 | 1.11 (0.10) | 1 |
| Biological Comparisons | # of Genes Tested | DE Genes (log2FC > 1, FDR ≤ 0.05) | DE Genes (log2FC > 1, FDR ≤ 0.01) | ||
|---|---|---|---|---|---|
| Up | Down | Up | Down | ||
| HUNFS0 vs. NOr | 10,210 | 1270 | 979 | 973 | 631 |
| HUNFs4 vs. HUNFS0 | 9714 | 407 | 935 | 181 | 668 |
| HUNFS4 vs. NOr | 10,054 | 944 | 789 | 816 | 477 |
| JCFS0 vs. NOr | 10,188 | 1400 | 860 | 1247 | 648 |
| JCFS4 vs. JCFS0 | 9605 | 497 | 1198 | 326 | 909 |
| JCFS4 vs. NOr | 10,096 | 1723 | 1677 | 1555 | 1291 |
| MERFS0 vs. NOr | 9840 | 346 | 348 | 224 | 201 |
| MERFS4 vs. MERFS0 | 6672 | 451 | 880 | 311 | 610 |
| MERFS4 vs. NOr | 9830 | 650 | 947 | 387 | 497 |
| Gene ID | Description | log2FC | FDR | Group |
|---|---|---|---|---|
| HUNFS4 vs. HUNFS0 | ||||
| AAEL007010 | CYP6AG4 | 2.86 | 5.93 × 10−6 | CYP |
| AAEL009117 | CYP6M5 | 1.52 | 8.47 × 10−4 | CYP |
| AAEL009762 | CYP307A1 | 2.06 | 0.0126 | CYP |
| JCFS4 vs. JCFS0 | ||||
| AAEL009762 | CYP307A1 | 3.53 | 1.76 × 10−5 | CYP |
| MERFS4 vs. MERFS0 | ||||
| AAEL002467 | Ae-Aper50 | 5.52 | 3.29 × 10−5 | CP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contreras-Perera, Y.; Mackenzie-Impoinvil, L.; Derilus, D.; Lenhart, A.; Rodriguez-Sanchez, I.P.; Manrique-Saide, P.; Flores, A.E. Deltamethrin Selection Drives Transcriptomic Changes in Detoxification, Immune, and Cuticle Genes in Aedes aegypti. Trop. Med. Infect. Dis. 2025, 10, 171. https://doi.org/10.3390/tropicalmed10060171
Contreras-Perera Y, Mackenzie-Impoinvil L, Derilus D, Lenhart A, Rodriguez-Sanchez IP, Manrique-Saide P, Flores AE. Deltamethrin Selection Drives Transcriptomic Changes in Detoxification, Immune, and Cuticle Genes in Aedes aegypti. Tropical Medicine and Infectious Disease. 2025; 10(6):171. https://doi.org/10.3390/tropicalmed10060171
Chicago/Turabian StyleContreras-Perera, Yamili, Lucy Mackenzie-Impoinvil, Dieunel Derilus, Audrey Lenhart, Iram P. Rodriguez-Sanchez, Pablo Manrique-Saide, and Adriana E. Flores. 2025. "Deltamethrin Selection Drives Transcriptomic Changes in Detoxification, Immune, and Cuticle Genes in Aedes aegypti" Tropical Medicine and Infectious Disease 10, no. 6: 171. https://doi.org/10.3390/tropicalmed10060171
APA StyleContreras-Perera, Y., Mackenzie-Impoinvil, L., Derilus, D., Lenhart, A., Rodriguez-Sanchez, I. P., Manrique-Saide, P., & Flores, A. E. (2025). Deltamethrin Selection Drives Transcriptomic Changes in Detoxification, Immune, and Cuticle Genes in Aedes aegypti. Tropical Medicine and Infectious Disease, 10(6), 171. https://doi.org/10.3390/tropicalmed10060171







