Non-O1, Non-O139 Vibrio cholerae Bacteremic Skin Infection with Multiple Skin Necrosis: Case Report
Abstract
1. Introduction
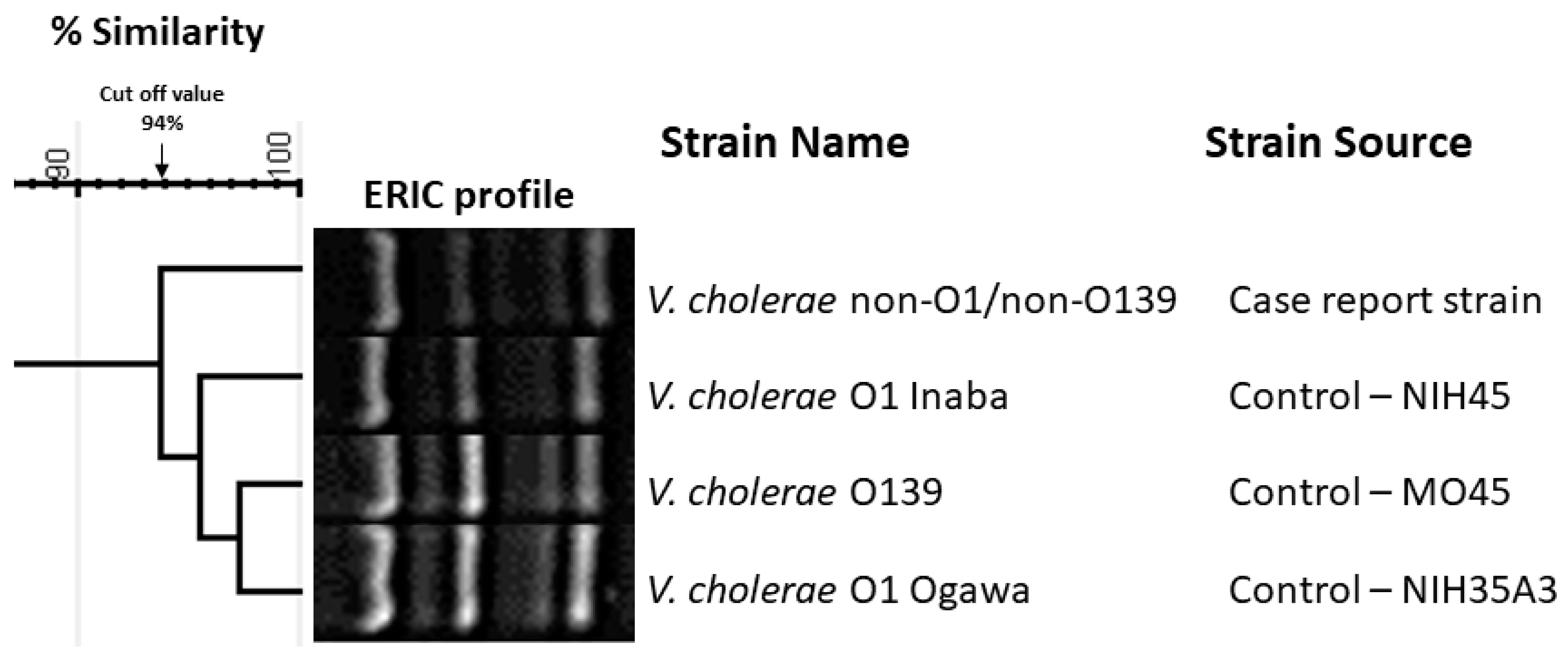
2. Case Presentations
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waldor, M.K.; Mekalanos, J.J. Lysogenic Conversion by a Filamentous Phage Encoding Cholera Toxin. Science 1996, 272, 1910–1914. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, T.; Mutreja, A.; Weill, F.X.; Das, B.; Ghosh, A.; Nair, G.B. Revisiting the Global Epidemiology of Cholera in Conjuction with the Genomics of Vibrio cholerae. Front. Public Health 2019, 7, 203. [Google Scholar] [CrossRef]
- Yamai, S.; Okitsu, T.; Shimada, T.; Katsube, Y. Distribution of Serogroups of Vibrio cholerae Non-O1 Non-O139 with Specific Reference to Their Ability to Produce Cholera Toxin, and Addition of Novel Serogroups. Kansenshogaku Zasshi. 1997, 71, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.V.; Matte, M.H.; Matté, G.R.; Jiang, S.; Sabeena, F.; Shukla, B.N.; Sanyal, S.C.; Huq, A.; Colwell, R. Molecular Analysis of Vibrio cholerae O1, O139, Non-O1, and Non-O139 Strains: Clonal Relationships between Clinical and Environmental Isolates. Appl. Environ. Microbiol. 2001, 67, 910–921. [Google Scholar] [CrossRef]
- Zhang, Q.; Alter, T.; Fleischmann, S. Non-O1/Non-O139 Vibrio cholerae—An Underestimated Foodborne Pathogen? An Overview of Its Virulence Genes and Regulatory Systems Involved in Pathogenesis. Microorganisms 2024, 12, 818. [Google Scholar] [CrossRef]
- Watve, S.; Barrasso, K.; Jung, S.A.; Davis, K.J.; Hawver, L.A.; Khataokar, A.; Palaganas, R.G.; Neiditch, M.B.; Perez, L.J.; Ng, W.-L. Ethanolamine Regulates CqsR Quorum-Sensing Signaling in Vibrio cholerae. BioRxiv 2019, 589390. [Google Scholar] [CrossRef]
- Krysenko, S.; Wohlleben, W. Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity. Med. Sci. 2022, 10, 40. [Google Scholar] [CrossRef]
- Ramamurthy, T.; Nandy, R.K.; Mukhopadhyay, A.K.; Dutta, S.; Mutreja, A.; Okamoto, K.; Miyoshi, S.-I.; Nair, G.B.; Ghosh, A. Virulence Regulation and Innate Host Response in the Pathogenicity of Vibrio cholerae. Front. Cell. Infect. Microbiol. 2020, 10, 572096. [Google Scholar] [CrossRef]
- Pitrak, D.L.; Gindorf, J.D. Bacteremic Cellulitis Caused by Non-Serogroup O1 Vibrio cholerae Acquired in a Freshwater Inland Lake. J. Clin. Microbiol. 1989, 27, 2874–2876. [Google Scholar] [CrossRef]
- Dutta, D.; Chowdhury, G.; Pazhani, G.P.; Guin, S.; Dutta, S.; Ghosh, S.; Rajendran, K.; Nandy, R.K.; Mukhopadhyay, A.K.; Bhattacharya, M.K.; et al. Vibrio cholerae Non-O1, Non-O139 Serogroups and Cholera-like Diarrhea, Kolkata, India. Emerg. Infect. Dis. 2013, 19, 464–467. [Google Scholar] [CrossRef]
- Chowdhury, G.; Joshi, S.; Bhattacharya, S.; Sekar, U.; Birajdar, B.; Bhattacharyya, A.; Shinoda, S.; Ramamurthy, T. Extraintestinal Infections Caused by Non-Toxigenic Vibrio cholerae Non-O1/Non-O139. Front. Microbiol. 2016, 7, 144. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.Y.; Duarte, C.; Rodríguez, G.J.; Montaño, L.A.; Benítez-Peñuela, M.A.; Díaz, P.; López, O.; Álvarez-Moreno, C.A. Bacteremia by Non-O1/Non-O139 Vibrio cholerae: Case Description and Literature Review. Biomedica 2023, 43, 323–329. [Google Scholar] [CrossRef]
- Aguinaga, A.; Portillo, M.E.; Yuste, J.R.; del Pozo, J.L.; García-Tutor, E.; Pérez-Gracia, J.L.; Leiva, J. Non-O1 Vibrio cholerae Inguinal Skin and Soft Tissue Infection with Bullous Skin Lesions in a Patient with a Penis Squamous Cell Carcinoma. Ann. Clin. Microbiol. Antimicrob. 2009, 8, 2007–2010. [Google Scholar] [CrossRef]
- Van Bonn, S.M.; Schraven, S.P.; Schuldt, T.; Heimesaat, M.M.; Mlynski, R.; Warnke, P.C. Chronic Otitis Media Following Infection by Non-01/Non-0139 Vibrio cholerae: A Case Report and Review of the Literature. Eur. J. Microbiol. Immunol. 2020, 10, 186–191. [Google Scholar] [CrossRef]
- Froelich, B.A.; Daines, D.A. In Hot Water: Effects of Climate Change on Vibrio–Human Interactions. Environ. Microbiol. 2020, 22, 4101–4111. [Google Scholar] [CrossRef]
- Deshayes, S.; Daurel, C.; Cattoir, V.; Parienti, J.J.; Quilici, M.L.; de La Blanchardière, A. Non-O1, Non-O139 Vibrio cholerae Bacteraemia: Case Report and Literature Review. Springerplus 2015, 4, 575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xiao, L.; Shan, X.; Dai, B.; Tang, C.; Xian, J.; Yu, Y. Case Report: Detection of Non-O1/Non-O139 Vibrio cholerae in a Patient with Hepatic Space-Occupying Lesions Using Metagenomic Next-Generation Sequencing. Front. Med. 2024, 11, 1483027. [Google Scholar] [CrossRef] [PubMed]
- Vithiya, G.; Velvizhi, S.; Sundaram, P.S.; Mandal, J. Non O1 Non O139 Vibrio cholerae Septicemia in a Patient with Carcinoma Pancreas-a Case Report and Literature Review, 2020–2023. Indian J. Med. Microbiol. 2024, 49, 100611. [Google Scholar] [CrossRef]
- Alex, V.; Moodley, M. Vibrio cholerae Bacteraemia: A Case Report on an Unusual Presentation. Diagn. Microbiol. Infect. Dis. 2025, 111, 116685. [Google Scholar] [CrossRef]
- Marino, A.; Cacopardo, B.; Villa, L.; D’Emilio, A.; Piro, S.; Nunnari, G. Think Vibrio, Think Rare: Non-O1-Non-O139-Vibrio cholerae Bacteremia in Advanced Lung Cancer—A Case Report. Trop. Med. Infect. Dis. 2024, 9, 224. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, Y.; Qian, H.; Liu, G.; Mei, Y.; Jin, F.; Xia, W.; Ni, F. Non-O1, Non-O139 Vibrio cholerae (NOVC) Bacteremia: Case Report and Literature Review, 2015–2019. Infect. Drug Resist. 2020, 13, 1009. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of Repetitive DNA Sequences in Eubacteria and Application to Finerpriting of Bacterial Enomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
- Miller, V.L.; Taylor, R.K.; Mekalanos, J.J. Cholera Toxin Transcriptional Activator ToxR Is a Transmembrane DNA Binding Protein. Cell 1987, 48, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Rivera, I.N.G.; Chun, J.; Huq, A.; Sack, R.B.; Colwell, R.R. Genotypes Associated with Virulence in Environmental Isolates of Vibrio cholerae. Appl. Environ. Microbiol. 2001, 67, 2421–2429. [Google Scholar] [CrossRef]
- Shirai, H.; Nishibuchi, M.; Ramamurthy, T.; Bhattacharya, S.K.; Pal, S.C.; Takeda, Y. Polymerase Chain Reaction for Detection of the Cholera Enterotoxin Operon of Vibrio cholerae. J. Clin. Microbiol. 1991, 29, 2517–2521. [Google Scholar] [CrossRef] [PubMed]
- Keasler, S.; Hall, R. Detecting and Biotyping Vibrio cholerae O1 with Multiplex Polymerase Chain Reaction. Lancet 1993, 341, 1661. [Google Scholar] [CrossRef]
- Singh, D.V.; Isac, S.R.; Colwell, R. Development of a Hexaplex PCR Assay for Rapid Detection of Virulence and Regulatory Genes in Vibrio cholerae and Vibrio mimicus. J. Clin. Microbiol. 2002, 40, 4321–4324. [Google Scholar] [CrossRef]
- Montero, D.A.; Vidal, R.M.; Velasco, J.; George, S.; Lucero, Y.; Gómez, L.A.; Carreño, L.J.; García-Betancourt, R.; O’Ryan, M. Vibrio cholerae, Classification, Pathogenesis, Immune Response, and Trends in Vaccine Development. Front. Med. 2023, 10, 1155751. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. Infections. Nat. Rev. Dis. Prim. 2018, 4, 1–19. [Google Scholar] [CrossRef]
- Issa, H. A Case of O1 Vibrio Cholera Bacteremia and Primary Peritonitis in a Patient with Liver Cirrhosis. Gastroenterol. Res. 2009, 2, 358–360. [Google Scholar] [CrossRef][Green Version]
- Schmidt, K.; Scholz, H.C.; Appelt, S.; Michel, J.; Jacob, D.; Dupke, S. Virulence and Resistance Patterns of Vibrio cholerae Non-O1/Non-O139 Acquired in Germany and Other European Countries. Front. Microbiol. 2023, 14, 1282135. [Google Scholar] [CrossRef]
- Saravanan, V.; Sanath Kumar, H.; Karunasagar, I.; Karunasagar, I. Putative Virulence Genes of Vibrio cholerae from Seafoods and the Coastal Environment of Southwest India. Int. J. Food Microbiol. 2007, 119, 329–333. [Google Scholar] [CrossRef]
- Hasan, N.A.; Ceccarelli, D.; Grim, C.J.; Taviani, E.; Choi, J.; Sadique, A.; Alam, M.C.; Siddique, A.K.; Bradley Sack, R.; Huq, A.; et al. Distribution of Virulence Genes in Clinical and Environmental Vibrio cholerae Strains in Bangladesh. Appl. Environ. Microbiol. 2013, 79, 5782–5785. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, M.; Rathnayake, I.U.; Huygens, F.; Nguyen, S.; Heron, B.; Jennison, A.V. Genomic and Evolutionary Insights into Australian Toxigenic Vibrio cholerae O1 Strains. Microbiol. Spectr. 2023, 11, e03617-22. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.D.; Lai, L.J.; Hsu, W.H.; Huang, T.Y. Vibrio cholerae Non-O1—The First Reported Case of Keratitis in a Healthy Patient. BMC Infect. Dis. 2019, 19, 916. [Google Scholar] [CrossRef]
- Abdelhafiz, T.A.; Alnimr, A.M.; Alabduljabbar, A.M.; AlMuqallad, H.S.; Abdulmonem Alzarra, A.; Alrashed, H.N.; Aladwani, M.M.; Hakami, A.M. Non O1 Vibrio cholerae as a Cause of Bacteremic Lower Limb Cellulitis: A Case Report. Int. J. Surg. Case Rep. 2019, 64, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, E.; Ochi, S.; Mizuno, T.; Morita, D.; Morita, M.; Ohnishi, M.; Koley, H.; Dutta, M.; Chowdhury, G.; Mukhopadhyay, A.K. Virulence of Cholera Toxin Gene-Positive Vibrio cholerae Non-O1/Non-O139 Strains Isolated from Environmental Water in Kolkata, India. Front. Microbiol. 2021, 12, 726273. [Google Scholar] [CrossRef]
- Aljindan, R.; Allahham, R.; Alghamdi, R.; Alhabib, I.; Alnassri, S.; Alkhalifa, W.; Diab, A.; Alomar, A.; Yamani, L.; Elhadi, N. Isolation and Characterization of Cholera Toxin Gene-Positive Vibrio cholerae Non-O1/Non-O139 Isolated from Urinary Tract Infection: A Case Report. Infect. Drug Resist. 2024, 17, 1147–1152. [Google Scholar] [CrossRef]
- Msemburi, W.; Karlinsky, A.; Knutson, V.; Aleshin-Guendel, S.; Chatterji, S.; Wakefield, J. The WHO Estimates of Excess Mortality Associated with the COVID-19 Pandemic. Nature 2023, 613, 130–137. [Google Scholar] [CrossRef]
- Alswaidi, F.M.; Assiri, A.M.; Alhaqbani, H.H.; Alalawi, M.M. Characteristics and Outcome of COVID-19 Cases in Saudi Arabia: Review of Six-Months of Data (March–August 2020). Saudi Pharm. J. 2021, 29, 682–691. [Google Scholar] [CrossRef]
- Alenzi, K.A.; Al-malky, H.S.; Altebainawi, A.F.; Abushomi, H.Q.; Alatawi, F.O.; Atwadi, M.H.; Khobrani, M.A.; Almazrou, D.A.; Alrubeh, N.; Alsoliabi, Z.A.; et al. Health Economic Burden of COVID-19 in Saudi Arabia. Front. Public Health 2022, 10, 927494. [Google Scholar] [CrossRef]
- Arsenault, C.; Gage, A.; Kim, M.K.; Kapoor, N.R.; Akweongo, P.; Amponsah, F.; Aryal, A.; Asai, D.; Awoonor-Williams, J.K.; Ayele, W.; et al. COVID-19 and Resilience of Healthcare Systems in Ten Countries. Nat. Med. 2022, 28, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, D.; Daza, J.; Liscano, Y. Coinfections and Superinfections Associated with COVID-19 in Colombia: A Narrative Review. Medicina 2023, 59, 1336. [Google Scholar] [CrossRef]
- Scott, H.; Zahra, A.; Fernandes, R.; Fries, B.C.; Thode Jr, H.C.; Singer, A.J. Bacterial Infections and Death among Patients with COVID-19 versus Non COVID-19 Patients with Pneumonia. Am. J. Emerg. Med. 2022, 51, 1–5. [Google Scholar] [CrossRef]
- Alqahtani, A.; Alamer, E.; Mir, M.; Alasmari, A.; Alshahrani, M.M.; Asiri, M.; Ahmad, I.; Alhazmi, A.; Algaissi, A. Bacterial Coinfections Increase Mortality of Severely Ill COVID-19 Patients in Saudi Arabia. Int. J. Environ. Res. Public Health 2022, 19, 2424. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Reytor, D.; Jaña, V.; Pavez, L.; Navarrete, P.; García, K. Accessory Toxins of Vibrio Pathogens and Their Role in Epithelial Disruption during Infection. Front. Microbiol. 2018, 9, 2248. [Google Scholar] [CrossRef]
- Zhao, Y.; He, T.; Tu, B.; Mao, X.; Jiang, J.; Jiang, X.; Wang, F.; Wang, M.; Wang, Y.; Sun, H. Death in a Farmer with Underlying Diseases Carrying Vibrio cholerae Non-O1/Non-O139 Producing Zonula Occludens Toxin. Int. J. Infect. Dis. 2022, 120, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Nagamani, R.; Al Momen, H.A.M. Non O1 Vibrio Cholerae: An Emerging Pathogen in Blood?—A Review and Report of Cases from a Regional Laboratory at the Eastern Province in Saudi Arabia. MRIMS J. Health Sci. 2017, 5, 109. [Google Scholar] [CrossRef]
- Kaki, R.; El-Hossary, D.; Jiman-Fatani, A.; Al-Ghamdi, R. Non-O1/Non-O139 Vibrio cholerae Septicaemia in a Saudi Man: A Case Report. JMM Case Rep. 2017, 4, e005077. [Google Scholar] [CrossRef]




| Test | Result | Normal Range |
|---|---|---|
| CRP | 26.71 mg/dL | 0.1–0.5 mg/dL |
| Procalcitonin | 4.46 μg/mL | 0.063–0.7 μg/mL |
| WBC | 17.8 k/μL | 4.0–11.0 k/μL |
| Hgb | 10.6 g/dL | 12.0–16.0 g/dL |
| Plt | 444 k/μL | 140–450 k/μL |
| Random sugar | 363 mg/dL | 70–140 mg/dL |
| HbA1c | 9.6% | 4–6% |
| ESR | 120 mm/h | 0–20 mm/h |
| Target | Nucleotide Sequence (5′-3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|
| toxR | CGGGATCCATGTTCGGATTAGGACAC CGGGATCCTACTCACACACTTTGATGGC | 900 | [23] |
| ompU | ACGCTGACGGAATCAACCA AAG GCGGAAGTTTGGCTTGAAG TAG | 869 | [24] |
| ctxA | CTCAGACGGGATTTGTTAGGCACG TCTATCTCTGTAGCCCCTATTACG | 301 | [25] |
| zot | TCGCTTAACGATGGCGCGTTTT AACCCCGTTTCACTTCTACCCA | 947 | [4] |
| tcpA | ACCAAATGCAACGCCGAATGGAGC GAAGAAGTTTGTAAAAGAAGAACAC | 617 | [26] |
| ace | TAAGGATGTGCTTATGATG GACACCC CGTGATGAATAAAGATACT CATAGG | 316 | [27] |
| hlyA | GAGCCGGCATTCATCTGAAT CTCAGCGGGCTAATACGGTTTA | 481 | [4] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alomar, A.I.; Elhadi, N.; Yamani, L.Z.; Allahham, R.; Alghamdi, R.; Alhabib, I.; Diab, A.; Mahmoud, N.; AlDossary, B.; Almejhim, M.; et al. Non-O1, Non-O139 Vibrio cholerae Bacteremic Skin Infection with Multiple Skin Necrosis: Case Report. Trop. Med. Infect. Dis. 2025, 10, 110. https://doi.org/10.3390/tropicalmed10040110
Alomar AI, Elhadi N, Yamani LZ, Allahham R, Alghamdi R, Alhabib I, Diab A, Mahmoud N, AlDossary B, Almejhim M, et al. Non-O1, Non-O139 Vibrio cholerae Bacteremic Skin Infection with Multiple Skin Necrosis: Case Report. Tropical Medicine and Infectious Disease. 2025; 10(4):110. https://doi.org/10.3390/tropicalmed10040110
Chicago/Turabian StyleAlomar, Amer Ibrahim, Nasreldin Elhadi, Lamya Zohair Yamani, Reema Allahham, Rana Alghamdi, Ibrahim Alhabib, Asim Diab, Nehal Mahmoud, Bashayer AlDossary, Mariam Almejhim, and et al. 2025. "Non-O1, Non-O139 Vibrio cholerae Bacteremic Skin Infection with Multiple Skin Necrosis: Case Report" Tropical Medicine and Infectious Disease 10, no. 4: 110. https://doi.org/10.3390/tropicalmed10040110
APA StyleAlomar, A. I., Elhadi, N., Yamani, L. Z., Allahham, R., Alghamdi, R., Alhabib, I., Diab, A., Mahmoud, N., AlDossary, B., Almejhim, M., Al-Romihi, N., Aldehalan, F., & Jindan, R. A. (2025). Non-O1, Non-O139 Vibrio cholerae Bacteremic Skin Infection with Multiple Skin Necrosis: Case Report. Tropical Medicine and Infectious Disease, 10(4), 110. https://doi.org/10.3390/tropicalmed10040110






