Socioconnectomics: Connectomics Should Be Extended to Societies to Better Understand Evolutionary Processes
Abstract
1. Introduction
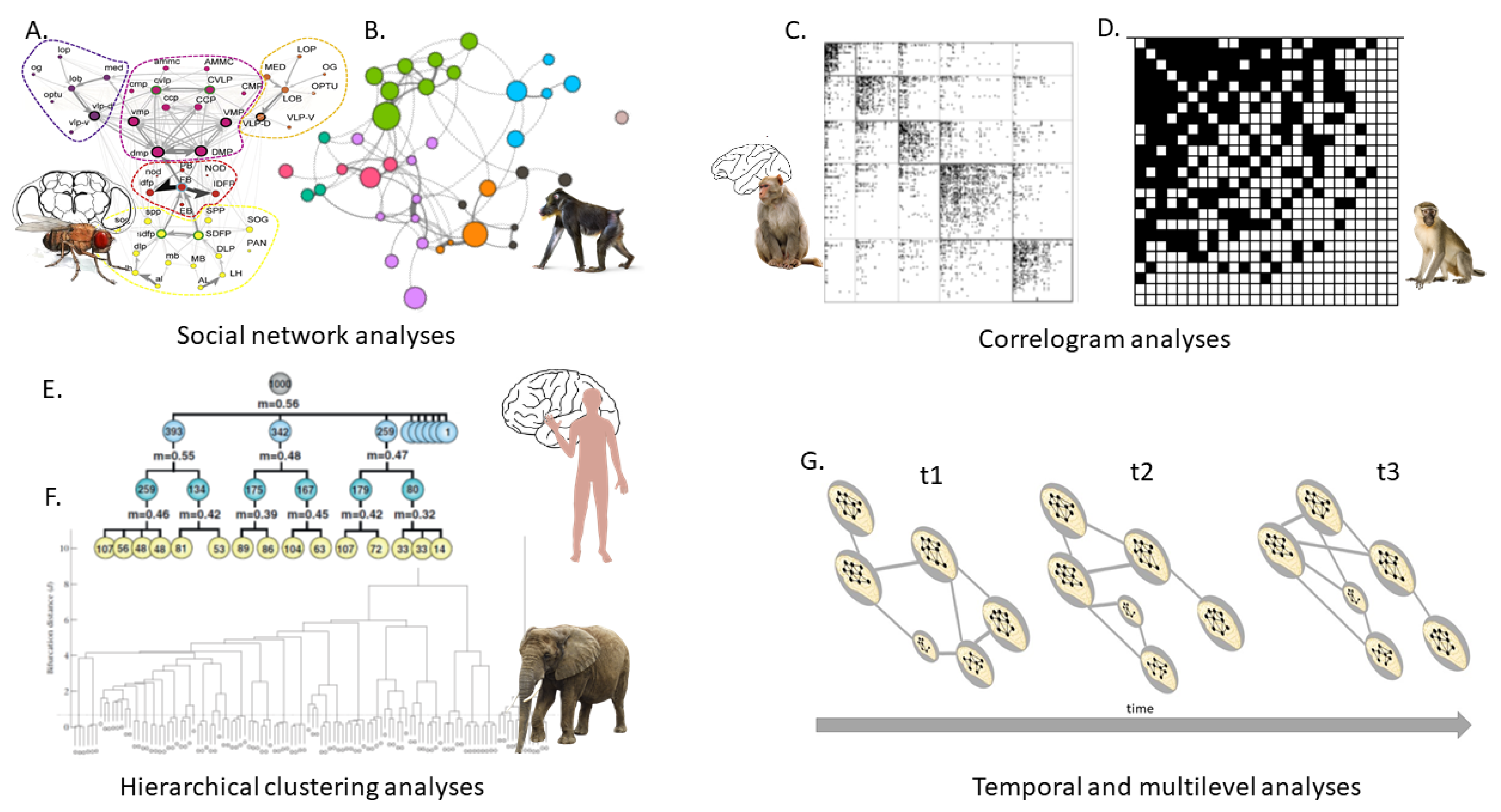
2. Universal Network Topological Properties
3. Network Analyses at Different Life Organizations
4. Same Challenges to Understanding Evolution
5. Future Research and Challenges
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morgan, J.L.; Lichtman, J.W. Why Not Connectomics? Nat. Methods 2013, 10, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, J.W.; Pfister, H.; Shavit, N. The Big Data Challenges of Connectomics. Nat. Neurosci. 2014, 17, 1448–1454. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.-N.; He, Y.; Betzel, R.F.; Colcombe, S.; Sporns, O.; Milham, M.P. Human Connectomics across the Life Span. Trends Cogn. Sci. 2017, 21, 32–45. [Google Scholar] [CrossRef] [PubMed]
- van den Heuvel, M.P.; Bullmore, E.T.; Sporns, O. Comparative Connectomics. Trends Cogn. Sci. 2016, 20, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Borgatti, S.P.; Mehra, A.; Brass, D.J.; Labianca, G. Network Analysis in the Social Sciences. Science 2009, 323, 892–895. [Google Scholar] [CrossRef] [PubMed]
- Sosa, S.; Sueur, C.; Puga-Gonzalez, I. Network Measures in Animal Social Network Analysis: Their Strengths, Limits, Interpretations and Uses. Methods Ecol. Evol. 2021, 12, 10–21. [Google Scholar] [CrossRef]
- Sueur, C.; Romano, V.; Sosa, S.; Puga-Gonzalez, I. Mechanisms of Network Evolution: A Focus on Socioecological Factors, Intermediary Mechanisms, and Selection Pressures. Primates 2019, 60, 167–181. [Google Scholar] [CrossRef]
- Fisher, D.; McAdam, A. Social Traits, Social Networks and Evolutionary Biology. J. Evol. Biol. 2017, 30, 2088–2103. [Google Scholar] [CrossRef]
- Dunbar, R. The Social Brain Hypothesis. Evol. Anthropol. Issues News Rev. 1998, 6, 178–190. [Google Scholar] [CrossRef]
- Muthukrishna, M.; Doebeli, M.; Chudek, M.; Henrich, J. The Cultural Brain Hypothesis: How Culture Drives Brain Expansion, Sociality, and Life History. PLoS Comput. Biol. 2018, 14, e1006504. [Google Scholar] [CrossRef]
- van Schaik, C.P.; Isler, K.; Burkart, J.M. Explaining Brain Size Variation: From Social to Cultural Brain. Trends Cogn. Sci. 2012, 16, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Oltvai, Z.N.; Barabási, A.-L. Life’s Complexity Pyramid. Science 2002, 298, 763–764. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Shen-Orr, S.; Itzkovitz, S.; Kashtan, N.; Chklovskii, D.; Alon, U. Network Motifs: Simple Building Blocks of Complex Networks. Science 2002, 298, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Avena-Koenigsberger, A.; Goñi, J.; Solé, R.; Sporns, O. Network Morphospace. J. R. Soc. Interface 2015, 12, 20140881. [Google Scholar] [CrossRef] [PubMed]
- Romano, V.; MacIntosh, A.J.J.; Sueur, C. Stemming the Flow: Information, Infection, and Social Evolution. Trends Ecol. Evol. 2020, 35, 849–853. [Google Scholar] [CrossRef]
- Silk, J.B.; Alberts, S.C.; Altmann, J. Social Bonds of Female Baboons Enhance Infant Survival. Science 2003, 302, 1231–1234. [Google Scholar] [CrossRef]
- McComb, K.; Moss, C.; Durant, S.M.; Baker, L.; Sayialel, S. Matriarchs As Repositories of Social Knowledge in African Elephants. Science 2001, 292, 491–494. [Google Scholar] [CrossRef]
- Nagy, M.; Akos, Z.; Biro, D.; Vicsek, T. Hierarchical Group Dynamics in Pigeon Flocks. Nature 2010, 464, 890–893. [Google Scholar] [CrossRef]
- Henrich, J. The Secret of Our Success: How Culture Is Driving Human Evolution, Domesticating Our Species, and Making Us Smarter; Princeton University Press: Princeton, NJ, USA, 2017; ISBN 0-691-17843-7. [Google Scholar]
- Farine, D.R.; Montiglio, P.O.; Spiegel, O. From Individuals to Groups and Back: The Evolutionary Implications of Group Phenotypic Composition. Trends Ecol. Evol. 2015, 30, 609–621. [Google Scholar] [CrossRef]
- Bijma, P. A General Definition of the Heritable Variation That Determines the Potential of a Population to Respond to Selection. Genetics 2011, 189, 1347–1359. [Google Scholar] [CrossRef]
- Bijma, P.; Wade, M. The Joint Effects of Kin, Multilevel Selection and Indirect Genetic Effects on Response to Genetic Selection. J. Evol. Biol. 2008, 21, 1175–1188. [Google Scholar] [CrossRef] [PubMed]
- Grampp, M.; Sueur, C.; van de Waal, E.; Botting, J. Social Attention Biases in Juvenile Wild Vervet Monkeys: Implications for Socialisation and Social Learning Processes. Primates 2019, 60, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Canteloup, C.; Hoppitt, W.; van de Waal, E. Wild Primates Copy Higher-Ranked Individuals in a Social Transmission Experiment. Nat. Commun. 2020, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Clune, J.; Mouret, J.B.; Lipson, H. The Evolutionary Origins of Modularity. Proc. Biol. Sci. 2013, 280, 20122863. [Google Scholar] [CrossRef]
- Bullmore, E.; Sporns, O. The Economy of Brain Network Organization. Nat. Rev. Neurosci. 2012, 13, 336–349. [Google Scholar] [CrossRef]
- Pasquaretta, C.; Levé, M.; Claidière, N.; van de Waal, E.; Whiten, A.; MacIntosh, A.J.J.; Pelé, M.; Bergstrom, M.L.; Borgeaud, C.; Brosnan, S.F.; et al. Social Networks in Primates: Smart and Tolerant Species Have More Efficient Networks. Sci. Rep. 2014, 4, 7600. [Google Scholar] [CrossRef]
- Romano, V.; Shen, M.; Pansanel, J.; MacIntosh, A.J.J.; Sueur, C. Social Transmission in Networks: Global Efficiency Peaks with Intermediate Levels of Modularity. Behav. Ecol. Sociobiol. 2018, 72, 154. [Google Scholar] [CrossRef]
- Romano, V.; Sueur, C.; MacIntosh, A.J.J. The Trade-off between Information and Pathogen Transmission in Animal Societies. Oikos 2022, 2022, e08290. [Google Scholar] [CrossRef]
- Thébault, E.; Fontaine, C. Stability of Ecological Communities and the Architecture of Mutualistic and Trophic Networks. Science 2010, 329, 853–856. [Google Scholar] [CrossRef]
- Strandburg-Peshkin, A.; Farine, D.R.; Couzin, I.D.; Crofoot, M.C. Shared Decision-Making Drives Collective Movement in Wild Baboons. Science 2015, 348, 1358–1361. [Google Scholar] [CrossRef]
- Sueur, C.; Jacobs, A.; Amblard, F.; Petit, O.; King, A.J. How Can Social Network Analysis Improve the Study of Primate Behavior? Am. J. Primatol. 2011, 73, 703–719. [Google Scholar] [CrossRef]
- Lusseau, D.; Conradt, L. The Emergence of Unshared Consensus Decisions in Bottlenose Dolphins. Behav. Ecol. Sociobiol. 2009, 63, 1067–1077. [Google Scholar] [CrossRef]
- Flack, J.C.; Girvan, M.; de Waal, F.B.; Krakauer, D.C. Policing Stabilizes Construction of Social Niches in Primates. Nature 2006, 439, 426–429. [Google Scholar] [CrossRef]
- Fruteau, C.; Voelkl, B.; van Damme, E.; Noë, R. Supply and Demand Determine the Market Value of Food Providers in Wild Vervet Monkeys. Proc. Natl. Acad. Sci. USA 2009, 106, 12007–12012. [Google Scholar] [CrossRef]
- Cantor, M.; Pires, M.M.; Marquitti, F.M.; Raimundo, R.L.; Sebastián-González, E.; Coltri, P.P.; Perez, S.I.; Barneche, D.R.; Brandt, D.Y.; Nunes, K.; et al. Nestedness across Biological Scales. PLoS ONE 2017, 12, e0171691. [Google Scholar] [CrossRef]
- Wittemyer, G.; Douglas-Hamilton, I.; Getz, W.M. The Socioecology of Elephants: Analysis of the Processes Creating Multitiered Social Structures. Anim. Behav. 2005, 69, 1357–1371. [Google Scholar] [CrossRef]
- Honey, C.J.; Sporns, O.; Cammoun, L.; Gigandet, X.; Thiran, J.P.; Meuli, R.; Hagmann, P. Predicting Human Resting-State Functional Connectivity from Structural Connectivity. Proc. Natl. Acad. Sci. USA 2009, 106, 2035–2040. [Google Scholar] [CrossRef]
- Dunbar, R.I.M. The Social Brain Meets Neuroimaging. Trends Cogn. Sci. 2012, 16, 101–102. [Google Scholar] [CrossRef]
- Sueur, C. Social Network, Information Flow and Decision-Making Efficiency: A Comparison of Humans and Animals. In Social Networking and Community Behavior Modeling; IGI Global: Hershey, PA, USA, 2011; pp. 164–177. [Google Scholar]
- Sueur, C.; King, A.J.; Pelé, M.; Petit, O. Fast and Accurate Decisions as a Result of Scale-Free Network Properties in Two Primate Species. In Proceedings of the Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2012; pp. 579–584. [Google Scholar]
- Marshall, J.A.R.; Bogacz, R.; Dornhaus, A.; Planqué, R.; Kovacs, T.; Franks, N.R. On Optimal Decision-Making in Brains and Social Insect Colonies. J. R. Soc. Interface 2009, 6, 1065–1074. [Google Scholar] [CrossRef]
- Bogacz, R. Optimal Decision-Making Theories: Linking Neurobiology with Behaviour. Trends Cogn. Sci. 2007, 11, 118–125. [Google Scholar] [CrossRef]
- Nowak, M.A. Five Rules for the Evolution of Cooperation. Science 2006, 314, 1560–1563. [Google Scholar] [CrossRef] [PubMed]
- Traulsen, A.; Nowak, M.A. Evolution of Cooperation by Multilevel Selection. Proc. Nat. Acad. Sci. USA 2006, 103, 10952–10955. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.A.; Tarnita, C.E.; Wilson, E.O. Nowak et al. Reply. Nature 2011, 471, E9–E10. [Google Scholar] [CrossRef]
- Leigh, E.G., Jr. The Group Selection Controversy. J. Evol. Biol. 2010, 23, 6–19. [Google Scholar] [CrossRef]
- Wilson, D.S. The Tide of Opinion on Group Selection Has Turned. In This View Life; Evolution Institute: Tampa, FL, USA, 2015; Volume 26. [Google Scholar]
- Wade, M.J. A Critical Review of the Models of Group Selection. Q. Rev. Biol. 1978, 53, 101–114. [Google Scholar] [CrossRef]
- Moscovice, L.R.; Sueur, C.; Aureli, F. How Socio-Ecological Factors Influence the Differentiation of Social Relationships: An Integrated Conceptual Framework. Biol. Lett. 2020, 16, 20200384. [Google Scholar] [CrossRef]
- Lesch, K.-P.; Bengel, D.; Heils, A.; Sabol, S.Z.; Greenberg, B.D.; Petri, S.; Benjamin, J.; Muller, C.R.; Hamer, D.H.; Murphy, D.L. Association of Anxiety-Related Traits with a Polymorphism in the Serotonin Transporter Gene Regulatory Region. Science 1996, 274, 1527–1531. [Google Scholar] [CrossRef]
- Wilson, A.J.; Gelin, U.; Perron, M.-C.; Réale, D. Indirect Genetic Effects and the Evolution of Aggression in a Vertebrate System. Proc. Biol. Sci. 2009, 276, 533–541. [Google Scholar] [CrossRef]
- Brent, L.J.N.; Heilbronner, S.R.; Horvath, J.E.; Gonzalez-Martinez, J.; Ruiz-Lambides, A.; Robinson, A.G.; Skene, J.H.P.; Platt, M.L. Genetic Origins of Social Networks in Rhesus Macaques. Sci. Rep. 2013, 3, srep01042. [Google Scholar] [CrossRef]
- Dawkins, R. The Selfish Gene: 30th Anniversary Edition; Oxford University Press: Oxford, UK, 2006; ISBN 978-0-19-157406-1. [Google Scholar]
- Whiten, A.; Ayala, F.J.; Feldman, M.W.; Laland, K.N. The Extension of Biology through Culture. Proc. Natl. Acad. Sci. USA 2017, 114, 7775–7781. [Google Scholar] [CrossRef]
- Nonacs, P.; Kapheim, K.M. Social Heterosis and the Maintenance of Genetic Diversity. J. Evol. Biol. 2007, 20, 2253–2265. [Google Scholar] [CrossRef] [PubMed]
- Finarelli, J.A.; Flynn, J.J. Brain-Size Evolution and Sociality in Carnivora. Proc. Natl. Acad. Sci. USA 2009, 106, 9345–9349. [Google Scholar] [CrossRef] [PubMed]
- DeCasien, A.R.; Williams, S.A.; Higham, J.P. Primate Brain Size Is Predicted by Diet but Not Sociality. Nat. Ecol. Evol. 2017, 1, 112. [Google Scholar] [CrossRef] [PubMed]
- Shultz, S.; Dunbar, R.I. Both Social and Ecological Factors Predict Ungulate Brain Size. Proc. Biol. Sci. 2005, 273, 207–215. [Google Scholar] [CrossRef]
- Burish, M.J.; Kueh, H.Y.; Wang, S.S. Brain Architecture and Social Complexity in Modern and Ancient Birds. Brain Behav. Evol. 2004, 63, 107–124. [Google Scholar] [CrossRef]
- Barrett, L.; Henzi, P.; Rendall, D. Social Brains, Simple Minds: Does Social Complexity Really Require Cognitive Complexity? Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 561–575. [Google Scholar] [CrossRef]
- Leadbeater, E.; Chittka, L. Social Learning in Insects—From Miniature Brains to Consensus Building. Curr. Biol. 2007, 17, R703–R713. [Google Scholar] [CrossRef]
- Sueur, C.; Quque, M.; Naud, A.; Bergouignan, A.; Criscuolo, F. Social capital: An independent dimension of healthy ageing. Peer Community J. 2021, 1, e23. [Google Scholar] [CrossRef]
- Almeling, L.; Hammerschmidt, K.; Sennhenn-Reulen, H.; Freund, A.M.; Fischer, J. Motivational Shifts in Aging Monkeys and the Origins of Social Selectivity. Curr. Biol. CB 2016, 26, 1744–1749. [Google Scholar] [CrossRef]
- Rosati, A.G.; Hagberg, L.; Enigk, D.K.; Otali, E.; Thompson, M.E.; Muller, M.N.; Wrangham, R.W.; Machanda, Z.P. Social Selectivity in Aging Wild Chimpanzees. Science 2020, 370, 473–476. [Google Scholar] [CrossRef]
- Albery, G.F.; Clutton-Brock, T.H.; Morris, A.; Morris, S.; Pemberton, J.M.; Nussey, D.H.; Firth, J.A. Ageing Red Deer Alter Their Spatial Behaviour and Become Less Social. Nat. Ecol. Evol. 2022, 6, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Testard, C.; Brent, L.J.N.; Andersson, J.; Chiou, K.L.; Negron-Del Valle, J.E.; DeCasien, A.R.; Acevedo-Ithier, A.; Stock, M.K.; Antón, S.C.; Gonzalez, O.; et al. Social Connections Predict Brain Structure in a Multidimensional Free-Ranging Primate Society. Sci. Adv. 2022, 8, eabl5794. [Google Scholar] [CrossRef] [PubMed]
- Solé, R.; Moses, M.; Forrest, S. Liquid Brains, Solid Brains. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190040. [Google Scholar] [CrossRef] [PubMed]
- Almaatouq, A.; Noriega-Campero, A.; Alotaibi, A.; Krafft, P.; Moussaid, M.; Pentland, A. Adaptive Social Networks Promote the Wisdom of Crowds. Proc. Natl. Acad. Sci. USA 2020, 117, 11379–113686. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sueur, C. Socioconnectomics: Connectomics Should Be Extended to Societies to Better Understand Evolutionary Processes. Sci 2023, 5, 5. https://doi.org/10.3390/sci5010005
Sueur C. Socioconnectomics: Connectomics Should Be Extended to Societies to Better Understand Evolutionary Processes. Sci. 2023; 5(1):5. https://doi.org/10.3390/sci5010005
Chicago/Turabian StyleSueur, Cédric. 2023. "Socioconnectomics: Connectomics Should Be Extended to Societies to Better Understand Evolutionary Processes" Sci 5, no. 1: 5. https://doi.org/10.3390/sci5010005
APA StyleSueur, C. (2023). Socioconnectomics: Connectomics Should Be Extended to Societies to Better Understand Evolutionary Processes. Sci, 5(1), 5. https://doi.org/10.3390/sci5010005





