4.2. Analysis
In line with the primary aims of this work, we started by assessing the different methods’ performance using the most widely used metric in the literature, namely the AUROC. Under this metric, the standard and the modified models stand on par with one another, the former achieving an AUROC value of 0.800 and the latter a marginally higher value of 0.806. We note that this is consistent with the previous reports in the literature, with the reported AUROC ranging from
(see Wang et al. [
5]) to
using the method proposed by Baltruschat et al. [
6]. The picture painted by comparing the per-label AUROC values, shown in
Table 2, is similar: on some labels, one model performs somewhat better, on others, the other does. Weighted by the frequencies of the labels, as we saw earlier, the difference all but disappears.
Both the standard and the modified model achieve nearly identical empirical AUROC scores, which, as we noted already, are normally used as the metric for ranking different methods in the field. Thus, superficially, this result suggests that the two methods are performing on par. Yet, in clinical terms, which is really what is of ultimate interest, this is far from the case: a closer look shows that the models actually perform rather differently.
Consider a slightly more nuanced comparison of the methods’ performances summarised in
Table 3. In terms of specificity and accuracy, the standard model can be seen to be superior. This is significant. For example, the difference of 0.023 in specificity means that out of 1000 patients, 23 can be (correctly) not subjected to further investigation and tests, thereby reducing the unnecessary discomfort caused and reducing the financial burden on the health care system. On the other hand, the modified model has a higher recall, so it is more likely to detect disease present in patients that have it. The difference in recall of 0.025 means it correctly diagnoses 25 more patients in 1000 than the standard model. To contextualise this, patients and healthcare professionals were willing to exchange 2250 FP diagnoses of colorectal cancer for one additional TP diagnosis [
23]. Similarly, 63% of women were found as >500 FPs reasonable per one life saved, and 37% would tolerate 10,000 or more [
24]. Some 1000 images have expert-drawn bounding boxes associated with them, localizing the visual presentation of the corresponding disease.
Reflecting on these observations, it is neither correct to say that the methods perform comparably, nor that one is superior to the other. Rather, there are significant differences between the two, and the question that is to be preferred in a specific context is one that demands collaborative consultative effort between teams of clinicians who understand the particular operative environment of interest and, no less importantly, medical ethicists whose role in the process is still inadequately appreciated.
4.3. Understanding the Data and the Interpretation of the Findings
A major concern of relevance to the efforts in the development of medical applications of machine learning concerns data used for training and testing algorithms. Notable problems include quality control (both of the data themselves, as well as their labelling), the clinical relevance and appropriateness of any associated annotations, the data balance, and numerous others. Indeed, concerns regarding the ChestX-ray14 corpus have been raised as well. Indeed, their nature mirrors the aforementioned pervasive ones: labelling accuracy (quality control), confounding information (quality control), clinical meaning of labels (quality control and clinical significance), and the usefulness of the labels (clinical significance and appropriateness) [
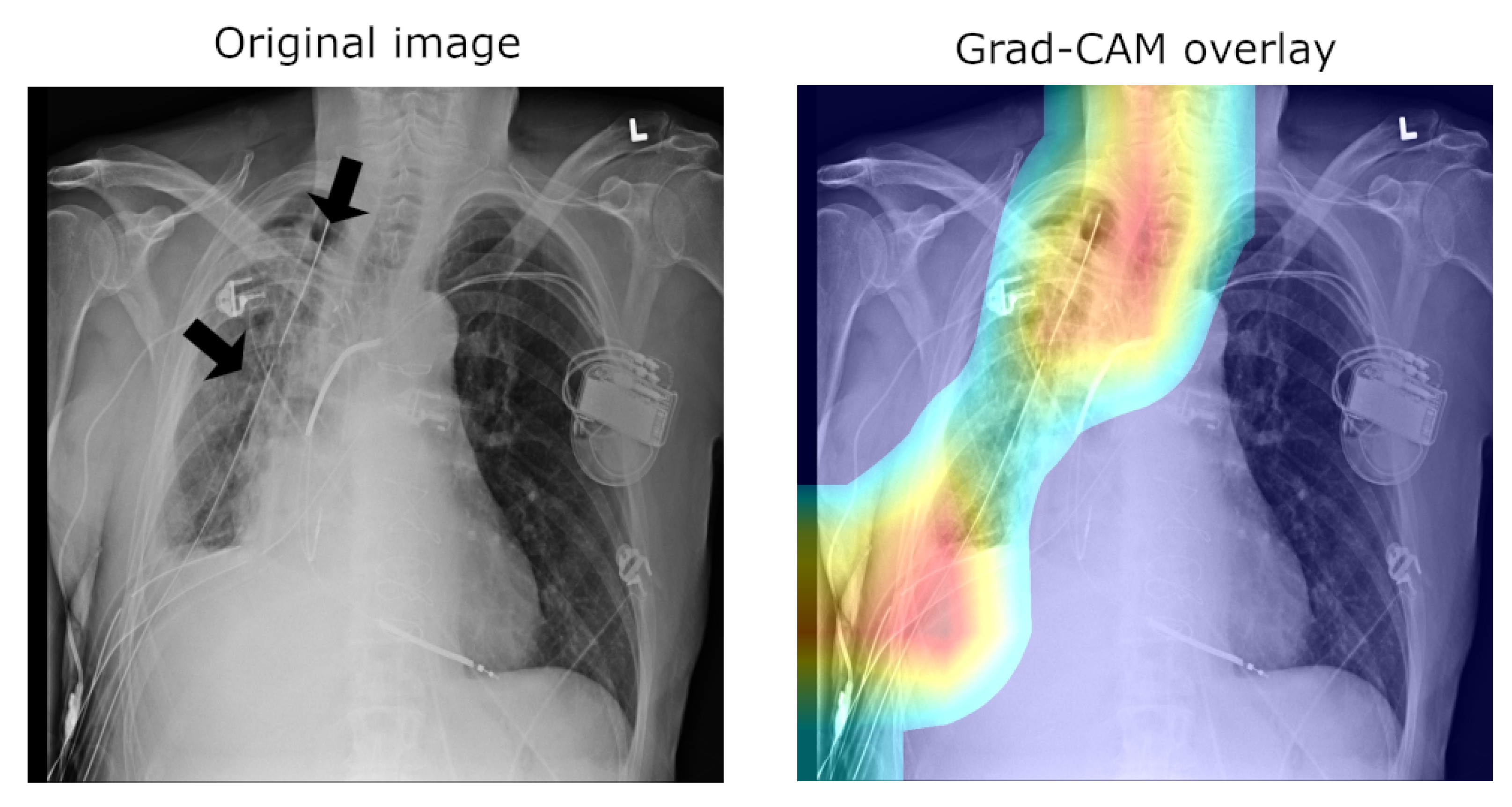
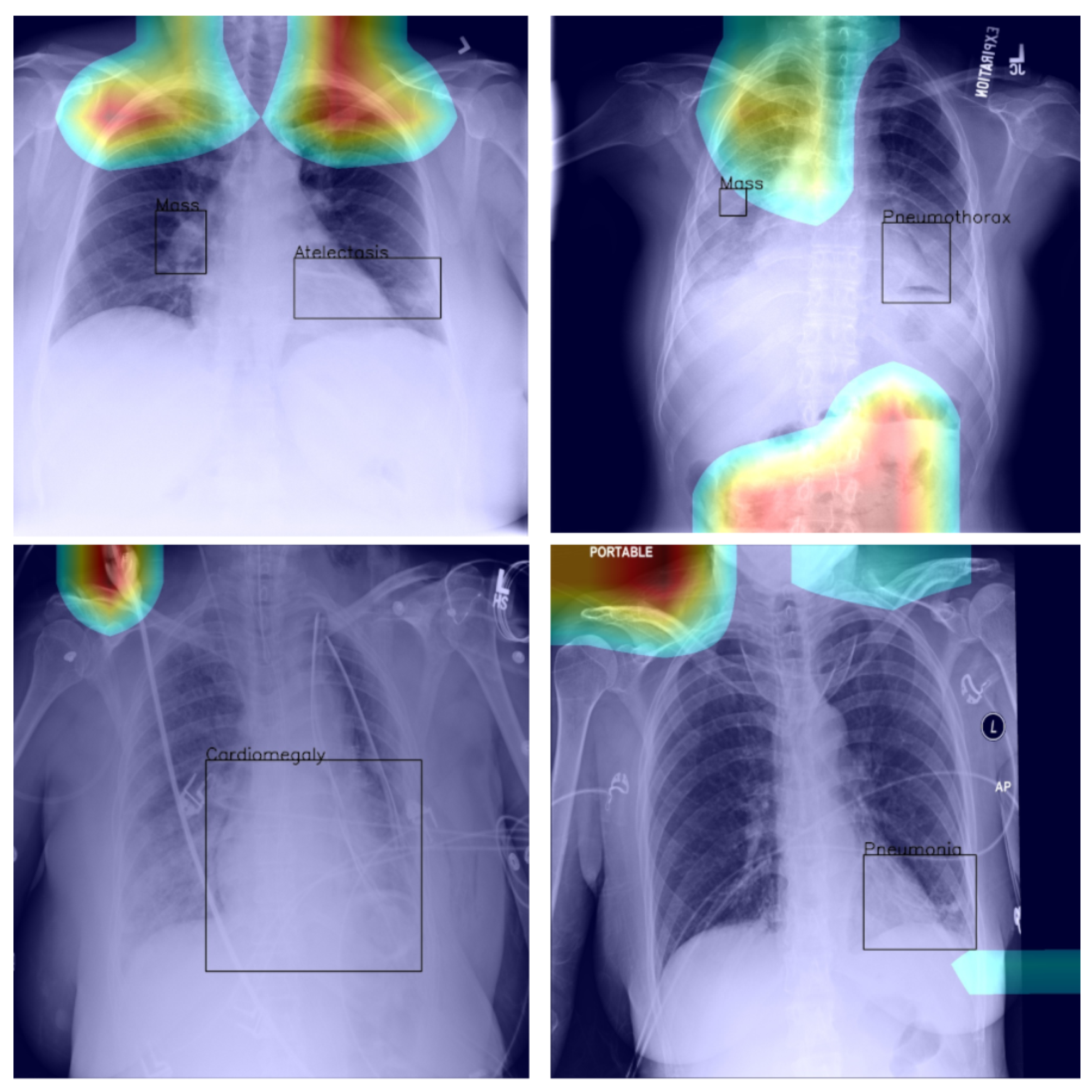
25]. Consider the following quality control concern: since some pneumothorax images are of patients that have already been treated and who hence have a chest drain, a machine-learning algorithm can learn to detect the presence of a drain and thus to correctly label the image, rather than learning to detect directly the condition itself (a similar issue in an anatomically different context was noted by Tun et al. [
26]). This is illustrated in
Figure 2, which shows on the left the original image, with the drain tube indicated, and on the right the learned class (pneumothorax) activation map.
Another important observation is that an image can have more than one class label associated with it (e.g., both the “Pneumonia” and “Infiltration” labels can be associated with the same X-ray image). Using the same loss function used to train the network, we can compute the mean model loss as a function of the number of labels,
N, associated with an image (n.b.
N ranges from zero for healthy lungs and goes up to eight, which is the maximum number of labels in this corpus). The loss increases at a linear rate with each additional label (see
Table 4), suggesting that the number of labels does not affect the per-label accuracy.
Looking at all instances of images with a single label and examining the mean activations across classes reveals a clear bias. An example is illustrated in
Table 5. The mean activation for the correct, ground truth label “Consolidation” is only 0.0842, whereas the mean activation for “Infiltration” is 0.2724—a 3.2-fold difference.
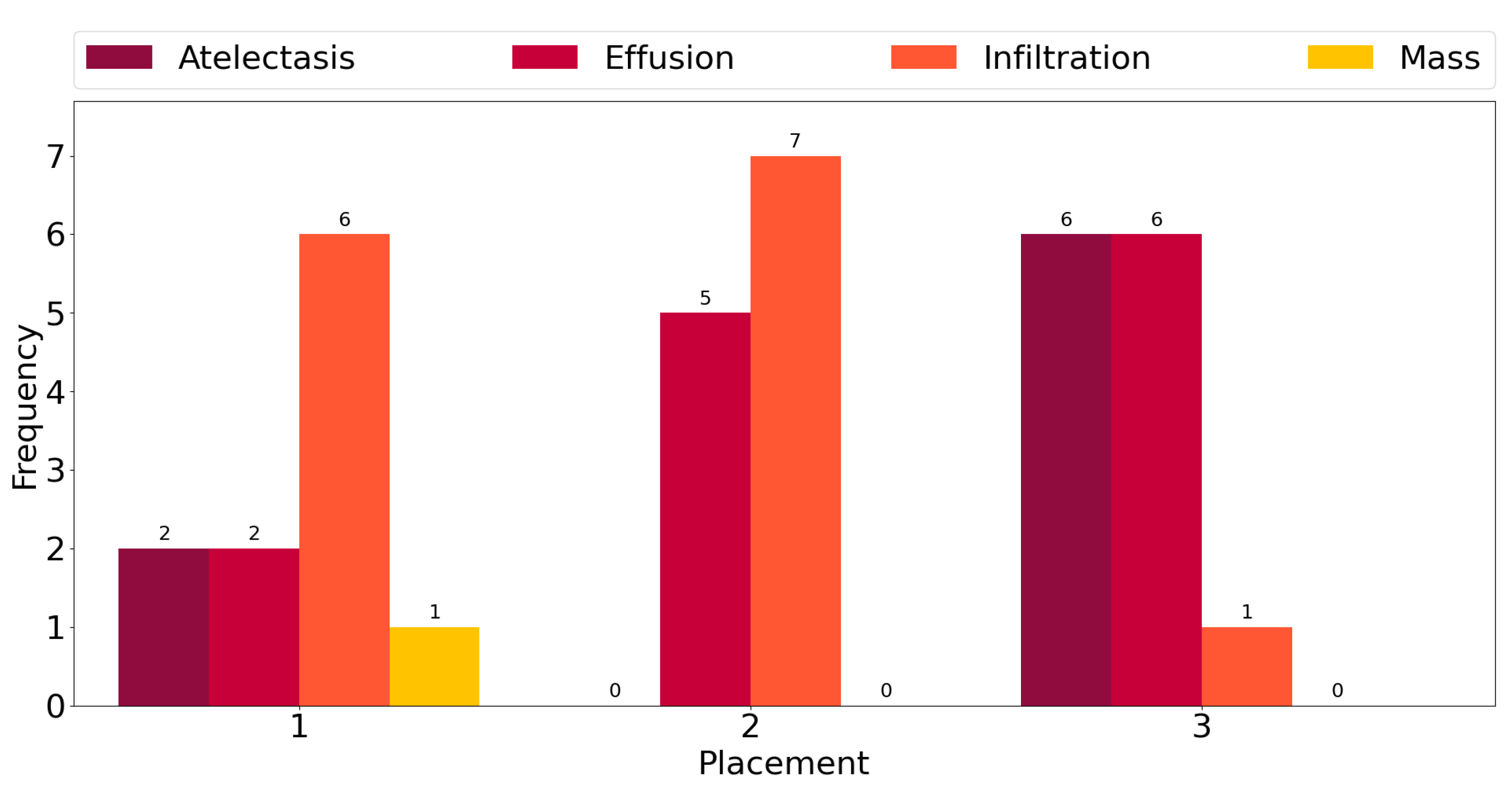
This observation is corroborated further by the plot in
Figure 3, which shows counts of the number of times each class exhibits among the three highest mean activations for single-label images across all classes. “Infiltration” is the most frequent class in the corpus, and for six out the fourteen ground truth labels, it exhibits the highest activation mean. In seven cases, it is the second-most-activated class, and in one, it is the third. In other words, it is
always amongst the top three most-activated output classes, regardless of what the true, target label is. The same can be seen for the three other most common classes, namely “Atelectasis”, “Effusion”, and “Mass”. The frequency of high activations is highly affected by the number of class instances in the corpus.
4.4. Saliency and Explainability
As we noted previously, using Grad-CAM, or indeed similar methods [
27], it is possible to quantify and thus visualise the importance of different parts of an analysed image a network uses as the basis for its prediction. This can be helpful both in understanding why the model fails when it does as well in focusing an expert’s attention for further analysis and interpretation.
In the context of the ChestX-ray8 corpus, the labelling within it has been criticised by some [
25]. This is hardly surprising, for the very manner in which the labels were extracted makes it impossible to consider them as the oracle ground truth. For instance, some pneumothorax images are of patients that have already had treatments indicated by a chest drains in the image. This can lead a network simply localising the drain and, based on this finding alone, label the image as belonging to the pneumothorax class, rather than as a result of the actual visual analysis of the presentation of the condition—see
Figure 2, wherein the drain has been marked with an arrow.
Using the bounding boxes information provided, we further examined how well the models performed in localizing the visual presentation of different diseases. We quantified this using the intersection over union (IoU), a ratio of the intersection of the bounding box and heat map activation and the union of the area they both cover, which can be loosely related to the well-known Bhattacharyya coefficient, a measurement of the amount of overlap between two statistical samples [
28,
29]. The ratio was calculated using thresholded heat maps with the key findings summarised in
Table 6 and
Table 7.
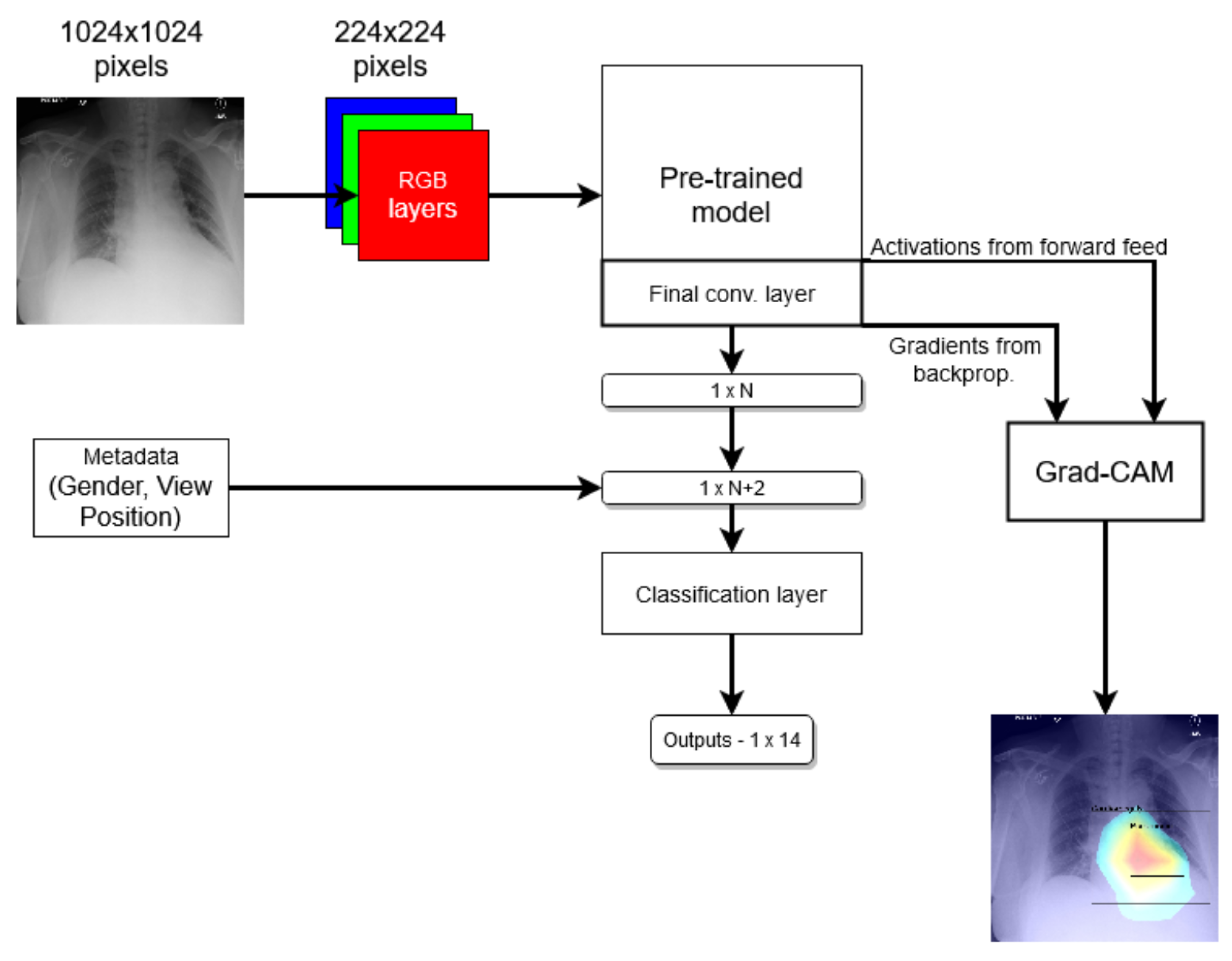
These findings are interesting in the context of the previously discussed AUROC-based comparison. Although the modified model performed better in terms of the former performance measure, that is to say AUROC, here, we found that it is the standard that does a better job in localizing the diseases. The likely explanation for this apparent paradox can be found in the structure of the network that was introduced in
Section 3 and shown in
Figure 1 and the flow of the metadata information and the manner in which it is used in the backpropagation. In any event, the important lesson to draw here is the same one that pervades the present article: any model must be examined in a variety of different ways and its performance measured using a range of comprehensive metrics and with a keen eye on their clinical significance, and its failure modes must be identified and understood, before any application in the real world is even considered.
Returning to the findings in
Table 6 and
Table 7, it is a concerning fact that the minimum IoU for
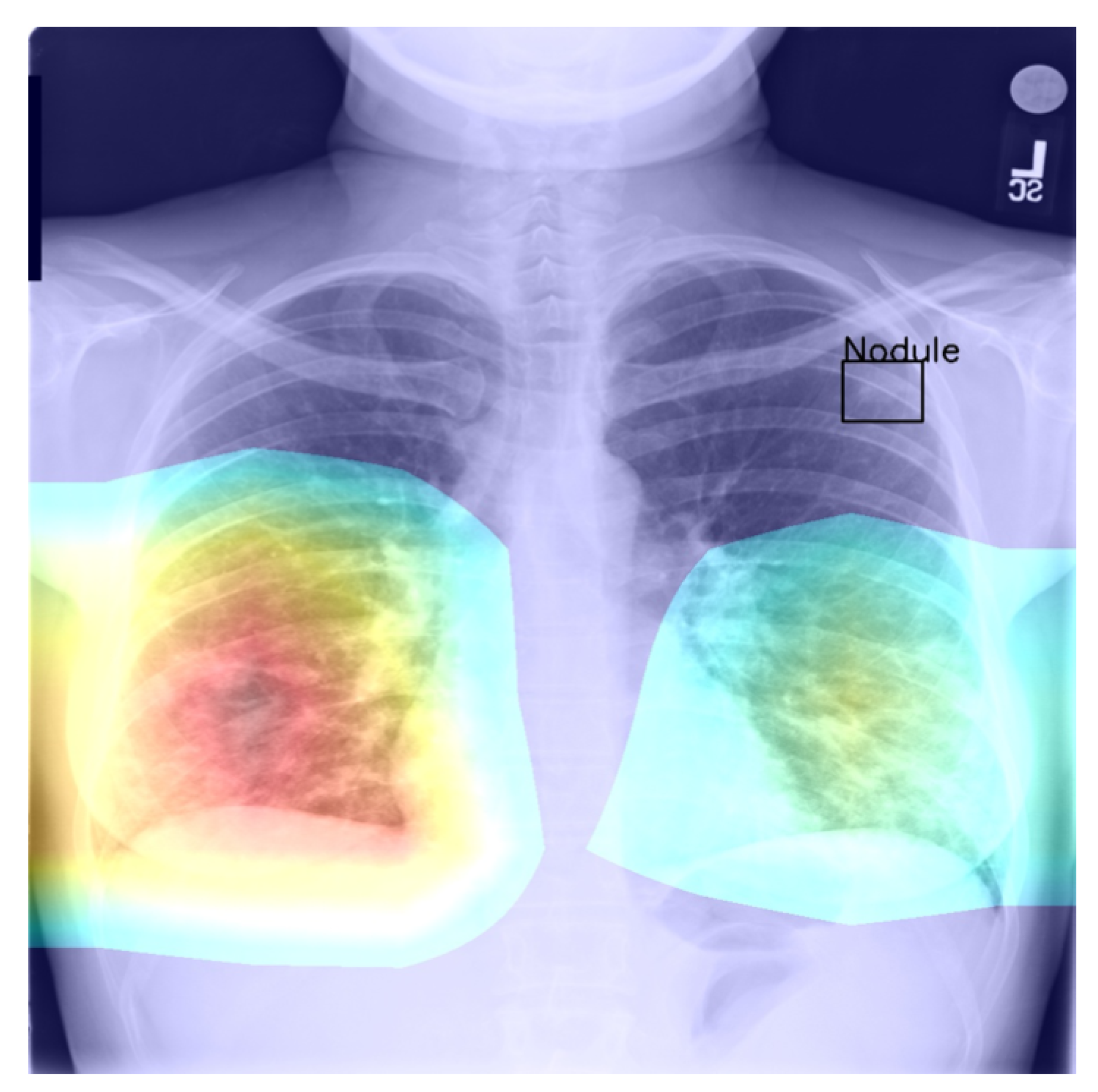
all classes was found to be zero (an interesting example is shown in
Figure 4. In other words, in the case of every class, that is disease, there was at least one instance in which the ground truth bounding box had no intersection with the thresholded saliency heat map. The models did, however, perform rather well in many cases, as indicated by the average scores. To check for potential biases, we measured the Pearson’s correlation coefficient between the number of class instances and the corresponding mean IoU and found it to be −0.265. This is an interesting and perhaps somewhat surprising finding, which should be revisited in future work. On the present evidence, we hypothesise that the more numerous classes exhibited greater variability in appearance presentation, which affected the performance under the IoU measure.
Last but not least, we found that when the heat map is significantly off course, this is often an indicator of there being confounding information—in the sense that it is not inherent information affected by imaging, but rather added by human experts such as radiologists and, as such, being assistive in correct labelling prediction, but misleading in the context of what the method is trying to achieve; see
Figure 5—present in an image (e.g., various textual descriptors overlaid on the images). If unnoticed, such information can instil false confidence in the performance of a model. Thus, we were again drawn to make two conclusions and recommendations: it is important that confounds of this kind be explicitly stated, observed, and discussed in any research and that a thorough examination of the data and specific findings be made whenever an algorithm is evaluated.
4.5. General Remarks
In this article, we focused specifically on the methodological assumptions underlying the prevalent approaches to the interpretation of deep-learning-based analysis of chest X-ray images. There are several reasons behind this choice. The most important of these lies in the practical importance of this application itself, a fact also reflected in the already large body of previous work, as discussed in
Section 1.
Secondly, the fact that the relevant phenomenology of the problem is well understood makes the desiderata and the potential pitfalls to watch for particularly clear, which is crucial for establishing a reliable and convincing framework needed for the challenging task of studying interpretability.
Notwithstanding the aforementioned focus, the applicability of the findings and the analysis we presented in this paper extend further and are not confined to the application of deep learning to chest X-ray image analysis. The issues we highlighted are readily identified in the use of deep learning in other medical applications, as well as non-medical ones. Perhaps the most fundamental question concerns the very premises of perturbation- and occlusion-based methods [
30,
31,
32,
33,
34,
35] to the determination of saliency and thus explainability, as recently also pointed out by Cooper et al. [
27].
Questions such as these are undoubtedly worth further study; meanwhile, caution should be exercised in the making of clinical conclusions based on insufficiently well-understood models.