1. Introduction
Cancer has long been acknowledged as a perilous disease with the potential for fatal outcomes. Lung cancer, a prevalent malignancy globally, stands out as a significant contributor to cancer-related mortality in both developed and developing nations. The majority of cases involve non-small-cell lung cancer (NSCLC), boasting a modest 5-year mortality rate of only 18%. Despite notable advancements in medical science leading to increased overall cancer survival rates, such progress is less pronounced in lung cancer due to the prevalence of advanced-stage cases among patients [
1]. Cancer cells typically migrate from the lungs to the lymph glands and then enter the bloodstream, with natural lymph flow directing the spread toward the chest’s center. Timely identification becomes crucial to preventing metastasis if the cancer spreads to other organs. Late-stage lesions are commonly treated with nonsurgical approaches like radiation, chemotherapy, surgical intervention, or monoclonal antibodies. This underscores the pivotal role of follow-up radiography in monitoring treatment response and tracking temporal changes in tumor radiography [
2]. Cancer analysis is usually conducted in a pathological laboratory using various methods. Microscopic examinations, including biopsies, and electronic modalities such as CT scans, ultrasound, and others are employed to examine cancerous tissue. Among these, the CT scan is the most commonly utilized pathological test and is highly favored for diagnosis. This imaging technique captures high-resolution, high-contrast images of the lungs from different perspectives, offering a three-dimensional assessment of the lesion.
Recent studies have introduced predictive algorithms that leverage the differential expression of genes to categorize lung cancer patients according to different health outcomes, including the likelihood of relapse and overall survival rates. Previous research has underscored the importance of biomarkers in treating non-small-cell lung cancer (NSCLC). The emergence of artificial intelligence (AI) has facilitated the quantitative evaluation of radiographic tumor features, an approach referred to as “radiomics” [
3]. Evidence from numerous studies suggests that non-invasive characterization of tumor features through radiomics offers enhanced predictive accuracy over traditional clinical evaluations.
Since the mid-19th century, pathologists have depended on traditional microscopy and glass slides to make precise diagnoses. This standard method requires pathologists to examine numerous glass slides by hand, a process that is both slow and requires significant effort. The advent of slide-scanning technology, which creates digital slides, has ushered classical pathology into a digital era, presenting various advantages for histopathology [
4]. A key benefit is the use of computer simulations, like automated image analysis, which aids medical professionals in examining and quantitatively evaluating slides. This advancement aims to reduce the duration of manual examinations and improve the accuracy, consistency, and efficiency of pathologists’ workflows. The employment of deep learning techniques for diagnostic support has recently sparked significant interest in histopathology.
In a clinical context, lung cancer CT scan images and normal lung images exhibit distinct characteristics that are crucial for accurate diagnosis. Radiologists analyze these images to identify potential abnormalities and distinguish between healthy and diseased lung tissue. For example, in CT scans, tumors or masses associated with lung cancer typically appear as areas of increased density on CT scans. They may be present as irregular, solid nodules or masses with varying degrees of density. However, normal lung tissue appears as a relatively homogenous pattern of lower density on CT scans. The texture is generally uniform, with no significant irregularities or masses.
For the effective classification of cancer and the selection of appropriate treatment options, a detailed analysis of lymph glands is crucial. Assessing multiple levels of lymph nodes is key for accurate prognosis and staging, which requires a thorough evaluation of lymph node condition [
5]. Recently, histopathological images have been identified as reliable indicators of various treatment biomarkers. However, the manual review of numerous slides is a demanding and time-consuming task for pathologists, who are prone to errors due to the challenge of remembering which sections have already been reviewed. Various solutions introduced in this field have been shown to outperform pathologists in terms of identifying micro-metastases with greater accuracy, especially under the pressure of a busy schedule. Similarly, in lung cancer, the presence of primary tumor metastases plays a crucial role in determining the stage of cancer, treatment possibilities, and patient outcomes, just as it does in breast cancer [
6].
DenseNet (Densely Connected Convolutional Networks) is a deep learning architecture [
7] that has demonstrated effectiveness in various image-related tasks, including medical image analysis. Its unique structure and characteristics contribute to its success in handling medical CT scans and histopathology images for cancer detection. The combination of DenseNet architecture with a Squeeze-and-Excitation (SE) block [
8], particularly for channel attention, enhances the model’s capabilities for medical image analysis. The SE block enhances DenseNet by introducing channel-wise attention. This allows the model to assign different levels of importance to different channels (features) in the intermediate representations. In medical images, where certain features or channels are more informative for detecting specific patterns associated with cancer, channel attention helps to focus on relevant information. Based on this idea, to surpass the accuracy levels of the existing deep-learning-based lung cancer detection methods, we propose an attention-based DenseNet (ATT-DenseNet) for lung cancer detection using CT scan images and histopathological images. We consider three baseline deep learning algorithms. First, we compare the performance of the proposed method with DenseNet having no attention mechanism. Then, we compare it with AlexNet [
9] followed by SqueezeNet [
10]. ATT-DenseNet outperforms these two baseline deep learning architectures by achieving increased accuracy and an increased F1-score. The novelty of this work as inspired by [
11] is summarized as follows:
We introduce the SE feature channel attention block into DenseNet architecture. This strategic incorporation aims to accentuate cancer-relevant information within the feature map. By dynamically recalibrating feature responses, our enhancement ensures greater emphasis on regions pertinent to cancer, thereby amplifying their significance in the overall analysis.
The network’s ability is enhanced to focus on crucial features by substituting average pooling with max pooling in the third transition layer. This modification strategically places additional emphasis on important regions during the analysis, improving the model’s capacity to capture relevant patterns associated with lung cancer.
Drawing inspiration from [
11], we implement an efficient method to prevent neuron deaths during model training. By addressing this challenge proactively, our methodology ensures the stability and robustness of the deep learning framework, leading to improved performance in lung cancer detection tasks.
Sophisticated data augmentation techniques are employed, including rotation, scaling, and horizontal flipping, to preprocess both CT scan images [
12] and histopathological images [
13]. Furthermore, normalization of the dataset is performed to maintain appropriate pixel value ranges, thereby preventing potential distortions caused by excessively high or low values. These preprocessing steps are crucial for enhancing the model’s ability to generalize across diverse datasets and improve overall performance.
In contrast with other algorithms, the ATT-DenseNet shows a notable boost in accuracy, precision, recall, and the F1-Score. It attains an average increase of 20% in accuracy, 19.66% in precision, 24.33% in recall, and 22.33% in the F1-Score across these performance measures.
In summary, the methodology represents a comprehensive and innovative approach to lung cancer detection, incorporating cutting-edge techniques in deep learning, strategic architectural enhancements, and meticulous data preprocessing strategies. By addressing key challenges and leveraging the latest advancements in the field, we aim to significantly improve the accuracy and reliability of lung cancer diagnosis, ultimately contributing to advancements in healthcare and patient outcomes.
The structure of the remainder of this paper is organized as follows:
Section 2 delves into related works, offering a concise review of current methodologies for detecting lung cancer. Following this,
Section 3 thoroughly describes the methodology employed, including details of the dataset used, the DenseNet architecture, and the implementation of the attention block.
Section 4 presents the experimental outcomes, showcasing a performance comparison with leading deep learning architectures currently considered state of the art. Finally,
Section 5 concludes the paper.
3. Materials and Methods
In this section, we first talk about the datasets that we use for lung cancer detection, consisting of chest CT scan images and histopathological images. Next, we discuss some details about the channel attention squeeze and excitation block, which we refer to as an SE block throughout this paper. Lastly, details regarding the DenseNet architecture are presented at the end of this section.
3.1. Dataset Description
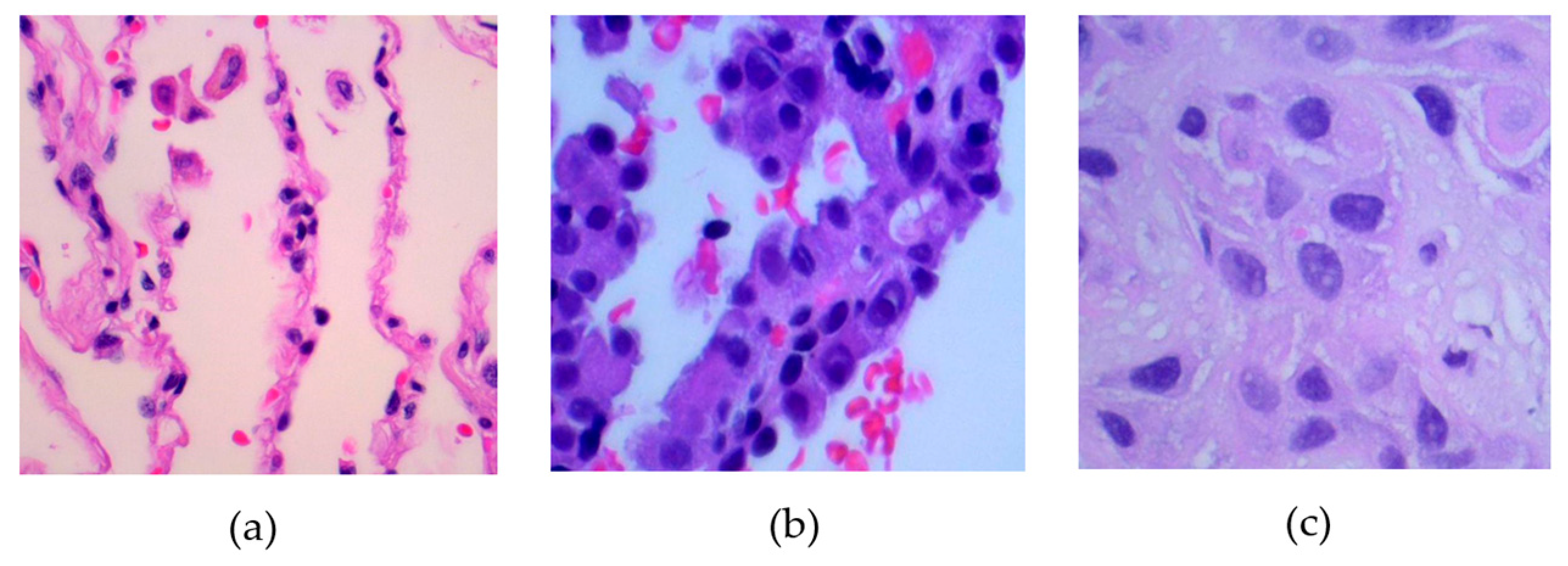
The dataset contains 15,000 histopathological images, each with dimensions of 768 × 768 pixels and stored in a JPEG format. These images come from a source that is compliant with HIPAA regulations and has been validated for accuracy. Initially, the collection included 750 images of lung tissue, which was equally divided among 250 images of benign lung tissue, 250 images of lung adenocarcinomas, and 250 images of lung squamous cell carcinomas.
Figure 1 presents samples associated with these three classes.
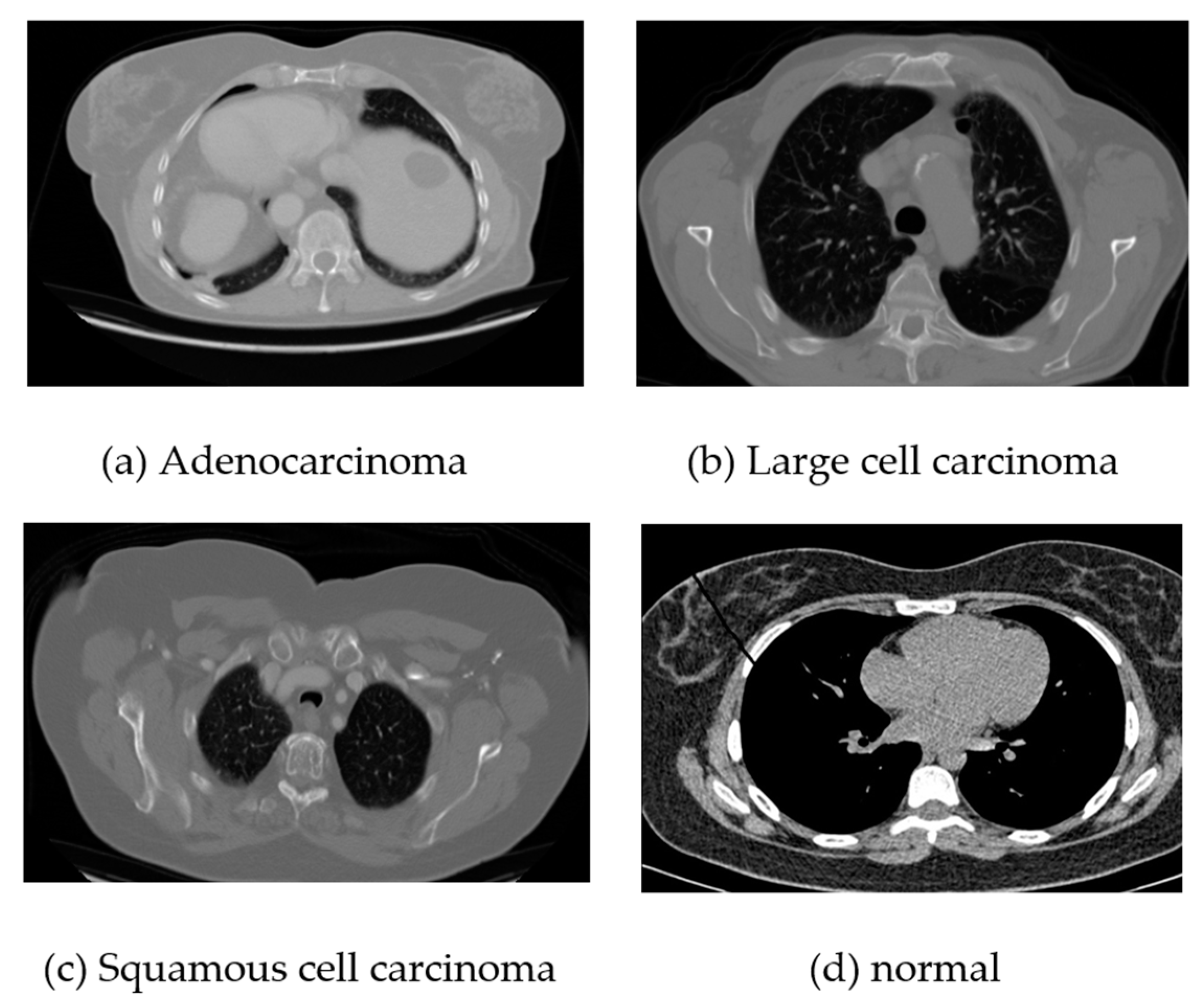
For the chest CT scan images, we consider three classes of cancer and a normal class. Among the cancer classes, there are three types: adenocarcinoma, large-cell carcinoma, and squamous cell carcinoma. A total of 340 images for each class including the normal class have been used for training and testing.
Figure 2 presents sample images from these classes.
3.2. DenseNet
DenseNet addresses significant challenges such as vanishing gradients and the inefficient use of parameters that are common in deep learning models [
31]. Its standout feature is the implementation of dense connectivity, ensuring that each layer is directly linked to every other layer in a feedforward fashion. This structure promotes the seamless flow of information and gradients across the network, enhancing learning efficiency and stability. The fundamental component of DenseNet is the dense block, which consists of a series of layers where each layer receives input from all preceding layers and, in turn, passes its feature maps forward. This pattern of dense connections fosters substantial feature reuse across the network, effectively leveraging and amalgamating learned representations from various depths. Moreover, DenseNet mitigates the issue of vanishing gradients by introducing shortcut connections that facilitate a more straightforward flow of gradients during the training process, thereby improving the model’s training efficiency and performance.
To manage the model’s complexity and computational cost, DenseNet incorporates transition layers between dense blocks. These transition layers typically include a combination of convolutional and pooling operations, serving to reduce the number of channels and downsample spatial dimensions. The growth rate, a hyperparameter, determines the number of additional channels introduced by each layer in the dense block, influencing the network’s capacity to learn and represent features.
In addition to dense connectivity, DenseNet often employs bottleneck layers within each dense block. These bottleneck layers consist of a 1 × 1 convolution followed by a 3 × 3 convolution, enhancing computational efficiency. The architecture typically concludes with a global average pooling layer, reducing spatial dimensions to a 1 × 1 grid, and a final classification layer.
The advantages of DenseNet include its ability to effectively utilize parameters, resulting in models with fewer parameters compared to traditional architectures. This efficient parameter usage, combined with dense connectivity, contributes to improved accuracy and training efficiency. DenseNet has proven particularly effective in image classification tasks and is widely adopted in the field of computer vision. A simplified representation of DenseNet is presented as follows:
Dense block: Assuming that input for the dense block is (output of the lth layer), is a set of convolutional operations. Next, a function is needed for concatenating input with the output of . The output of the lth layer is the concatenation of all previous layer outputs and the output of the operation.
Transition layer: Input,
(output of the
lth dense block).
is the compression factor that reduces the number of channels. Next, convolution and average pooling operations are performed. The output of the transition layer can be represented as follows:
The transition layer is used to reduce the number of channels and spatial dimensions, aiding in parameter efficiency.
Overall DenseNet Structure: If we assume as an input image, and , , …, are dense blocks, , , …, are compression factors for each transition layer. is the growth rate (number of additional channels added by each layer)
The overall structure is a sequence of dense blocks connected by transition layers, forming a dense and interconnected network. The DenseNet architecture for detecting lung cancer using chest CT scans and histopathological images is presented in
Figure 3.
3.3. SE Block
The attention mechanism functions as a mechanism for resource allocation and can be categorized into various types, including channel attention, pixel attention, multistage attention, and others. In this study, we focus on the channel attention SE block, as introduced in [
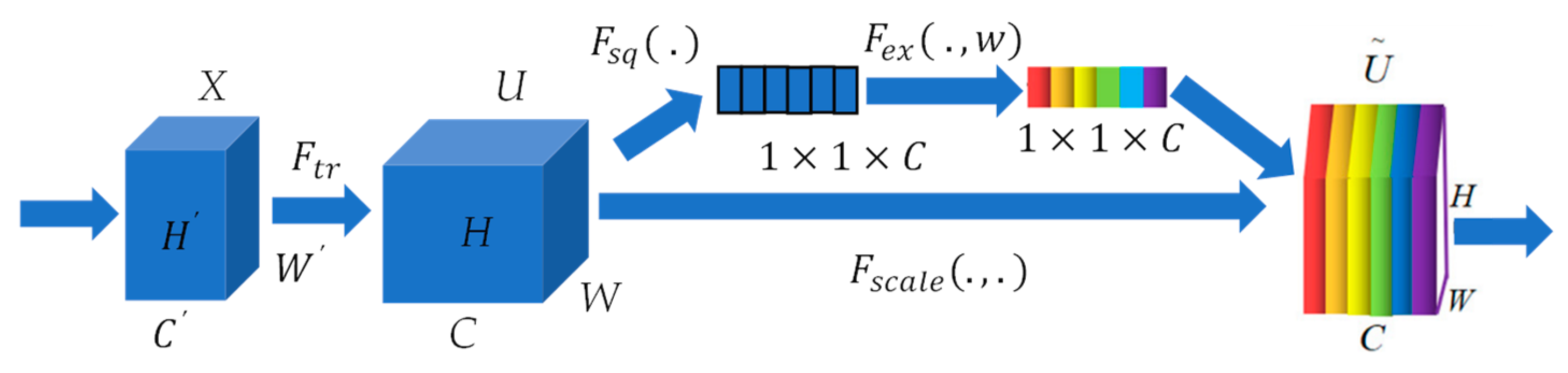
32]. The fundamental concept of this block is to determine feature weights based on the loss, assigning greater weight to effective feature maps. The SE block consists primarily of two components: squeeze and excitation.
The squeeze operation in neural network architectures compresses features along the spatial dimensions, effectively transforming each two-dimensional feature map into a single real number. This compression process generates a global receptive field, allowing the network to capture and respond to global information present in the input data. On the other hand, the excitation step is akin to the gating mechanism found in Recurrent Neural Networks (RNNs). This mechanism involves selectively amplifying or dampening specific features based on their relevance to the task at hand, enabling the network to focus on the most informative parts of the input data. Together, squeeze and excitation operations allow a neural network to dynamically adjust the importance of different channels, enhancing its ability to learn complex patterns and relationships in the data. It generates weights for each feature channel using parameters and learns these weights to explicitly model the correlation between feature channels. The graphical representation of the SE block is depicted in
Figure 4. For any given transformation
, mapping the input
to the feature map
where
, we can construct a corresponding SE block to perform feature recalibration. The feature
first passes through a squeeze operation
, which compresses
into a
feature. Next, through excitation operation
, the feature from
is excited. Lastly, through
, the recalibration feature is achieved, where
implies that the weights assigned to the excitation output are individually applied to each preceding feature channel through multiplication. Through this process, the original characteristics in the channel dimension are recalibrated, achieving a fine-tuning of the importance of each feature channel.
The ATT-DenseNet model leverages a sophisticated attention mechanism, specifically the squeeze-and-excitation (SE) block, to improve its performance in lung cancer detection using CT and histopathological images. Here is a detailed explanation of how this attention mechanism is implemented and its impact on the model’s performance:
3.3.1. Implementation of the SE Block
Squeeze Operation: This component of the SE block compresses the spatial dimensions of the feature maps. Each two-dimensional feature map is aggregated into a single real number, effectively summarizing the spatial information into a channel descriptor. This operation creates a global receptive field for each channel, capturing global spatial information succinctly.
Excitation Operation: The excitation step follows the squeezing of feature maps. It uses a fully connected layer to learn a set of weights for each channel. These weights are learned during training and are used to model the interdependencies between the channels. The operation is similar to a gating mechanism in recurrent neural networks (RNNs), allowing the model to assign adaptive importance to each channel based on the current input.
Feature Recalibration: The output from the excitation step, which consists of weights for each channel, is used to recalibrate the original feature maps. This is achieved by scaling each channel of the feature map by its corresponding learned weight. The recalibrated feature maps emphasize informative features while suppressing less useful ones, enhancing the representational power of the network.
3.3.2. Impact on Model Performance
Enhanced Feature Representation: By dynamically recalibrating the feature channels based on their relevance to the task at hand, ATT-DenseNet can focus more on important features while ignoring irrelevant ones. This results in a more discriminative feature representation, improving the model’s accuracy in detecting lung cancer.
Improved Model Generalization: The ability to adaptively adjust the importance of features based on the input allows ATT-DenseNet to generalize better across different datasets and imaging conditions. This adaptability is crucial in medical imaging, where variability across images is common.
Efficient Use of Model Parameters: Despite the added complexity of the attention mechanism, the SE block’s efficient design ensures that the increase in computational cost and parameters is minimal. This efficiency is particularly important in medical applications, where model deployment may need to be resource-conscious.
In summary, the attention mechanism used in ATT-DenseNet, embodied by the SE block, plays a critical role in enhancing the model’s ability to detect lung cancer from CT and histopathological images. By focusing on relevant features and suppressing irrelevant ones, the model achieves improved performance metrics, including accuracy, precision, recall, and the F1-score, making it a powerful tool for early and accurate lung cancer detection.
The SE block significantly enhances the functionality of the ATT-DenseNet model by introducing a novel approach to feature recalibration within deep learning architectures. In its functionality, the SE block first executes a squeeze phase, which aggregates spatial information across the feature maps by compressing the spatial dimensions of each channel into a single value. This process captures global contextual information from each feature map, providing a comprehensive summary of the spatial attributes. Following this, the excitation phase employs a fully connected layer to learn weights for each channel, effectively determining the importance of each feature based on the aggregated global information. This mechanism enables the model to understand the intricate relationships between channels and to assign more weight to those features deemed crucial for the task at hand. The culmination of the SE block’s process is the feature recalibration stage, wherein the original feature maps are scaled by the learned channel-specific weights. This recalibration allows the model to enhance or suppress features based on their learned importance, optimizing the network’s focus and resource allocation towards the most informative features for the specific task of lung cancer detection.
The role of the SE block in emphasizing cancer-related information within the feature maps is pivotal for the enhanced performance of the ATT-DenseNet model in detecting lung cancer. By dynamically adjusting the emphasis on different features, the SE block enables the model to concentrate more effectively on the characteristics indicative of cancerous tissues, such as abnormal growth patterns and irregular tissue structures, while diminishing the focus on less relevant features. This selective attention to cancer-related features allows the model to be more sensitive and accurate in identifying potential cancerous lesions within CT and histopathological images. The ability to discern and prioritize critical cancer-related information over normal tissue characteristics significantly improves the model’s diagnostic precision, enabling it to detect lung cancer with greater accuracy, precision, recall, and F1-score. Through the implementation of the SE block, ATT-DenseNet advances the field of medical imaging analysis by providing a more effective tool for the early detection and diagnosis of lung cancer, potentially leading to better patient outcomes through timely and accurate treatment planning.
3.4. Neuron Death Avoidance
PReLU (Parametric Rectified Linear Unit) is a variation of the popular ReLU (Rectified Linear Unit) activation function commonly used in neural networks [
33]. While ReLU sets all negative values to zero, PReLU allows for a small negative slope, which is learned during training. Neuron death, or dying ReLU problem, can occur when the ReLU units always output zero for a particular input during training, leading to those units no longer updating their weights. This can happen when the input to a ReLU neuron is consistently negative, causing the gradient to be zero and thus preventing the weights from being updated. By allowing for a small negative slope in PReLU, even for negative inputs, it helps to alleviate this issue by providing a non-zero gradient for those inputs. This encourages the flow of gradients during backpropagation, preventing neurons from becoming inactive during training.
3.5. Pseudocode of the Proposed Method
The pseudocode describes a step-by-step method to analyze medical images for lung cancer using a special computer program. It starts by preparing the images in a consistent way. Then, for each image, it goes through several layers that automatically extract important features. A special attention mechanism then focuses on the most relevant features, making them more prominent for the final decision-making process. Finally, the program decides whether the image shows signs of lung cancer. The effectiveness of this process is checked by seeing how accurate and reliable the decisions are.
Input: CT Scan and Histopathological Images;
Preprocessing: Normalize images to the same scale;
For each image in the dataset:
- a.
Pass image through DenseNet layers:
- -
Convolutional layers: Extract features from the image;
- -
Pooling layers: Reduce spatial dimensions of the feature maps;
- -
Dense blocks: Enhance feature extraction through densely connected layers;
- b.
Integrate Attention Mechanism:
- -
Compute attention scores for feature maps generated by DenseNet;
- -
Multiply attention scores with corresponding DenseNet feature maps;
- c.
Classification Layer:
- -
Flatten the attended feature maps;
- -
Pass through fully connected layers to obtain the final classification;
Output: Lung cancer diagnosis (Cancerous or Non-Cancerous);
Evaluation:
- -
Use metrics such as accuracy, precision, recall, and the F1-Score to evaluate model.
The problem statement in this paper focuses on the challenge of detecting lung cancer accurately and efficiently using CT and histopathological images. Despite advances in medical imaging, accurately diagnosing lung cancer early remains difficult due to the complex nature of tumor appearances and variations in imaging. This paper introduces the ATT-DenseNet model as a solution to improve diagnostic performance by leveraging deep learning and attention mechanisms to enhance the model’s ability to focus on relevant features within the images, aiming to increase the accuracy, precision, recall, and F1-score of lung cancer detection. In the medical industry, this model could significantly impact early lung cancer diagnosis, treatment planning, and patient outcomes. It could be integrated into diagnostic imaging systems in healthcare facilities to assist radiologists and pathologists, enhancing the precision of lung cancer detection and enabling more personalized treatment strategies, ultimately leading to better patient care and survival rates.
4. Results
4.1. Parameter Settings and Implementation Details
For all the experimental results conducted, we used AMD Ryzen 7-5800HS CPU, having 40 GB of randomly accessible memory. NVIDIA GeForce RTX 4060 GPU was used for simulation. Furthermore, we used Tensorflow 2.15.0 and Python 3.10.12 for all the results acquired. Implementation details of the proposed ATT-DenseNet are described in
Table 1. We used 70% of the data for training, and rest of the 30% data were used for validation.
4.2. Histopathological Images
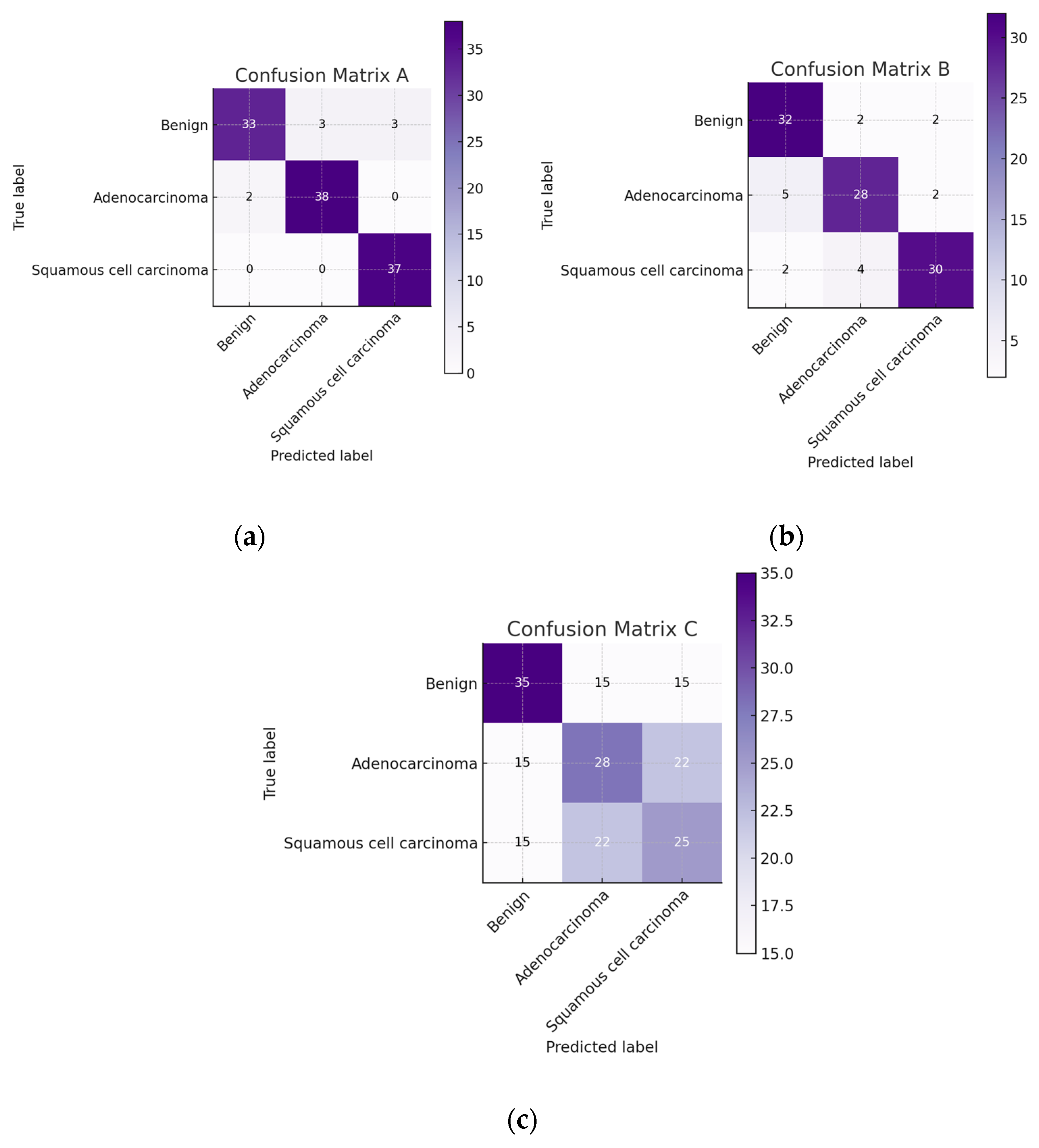
First, we present the results associated with cancer detection for histopathological images. In the context of evaluating the performance of classification models within machine learning, the confusion matrix stands out as a pivotal tool. This matrix provides a detailed breakdown of the predictions made by a model, allowing for a nuanced assessment of its performance in terms of accurately predicting different classes. The confusion matrix is structured as a table, categorizing predictions into four fundamental types: True Positives (TP), True Negatives (TN), False Positives (FP), and False Negatives (FN). TPs and TNs represent instances where the model has correctly predicted the positive and negative classes, respectively. By contrast, FPs and FNs reflect the errors in prediction, where the model incorrectly identifies the positive and negative classes.
Derived from the confusion matrix, several key performance metrics that offer insights into different aspects of the model’s predictive accuracy are accuracy, precision, recall, and F1 score. These metrics, derived from the confusion matrix, are instrumental in comprehensively understanding the performance of a classification model. They highlight not only the model’s accuracy but also the nature and extent of any prediction errors, thereby guiding further model refinement and optimization.
We had 750 images in total representing three different classes and an even distribution of 250 images for each class. We reserved 15% of the data for validation, and the results presented are based on this validation data performance.
Figure 5 presents the confusion matrix for ATT-DenseNet implementation along with two other baselines, namely AlexNet and SqueezeNet.
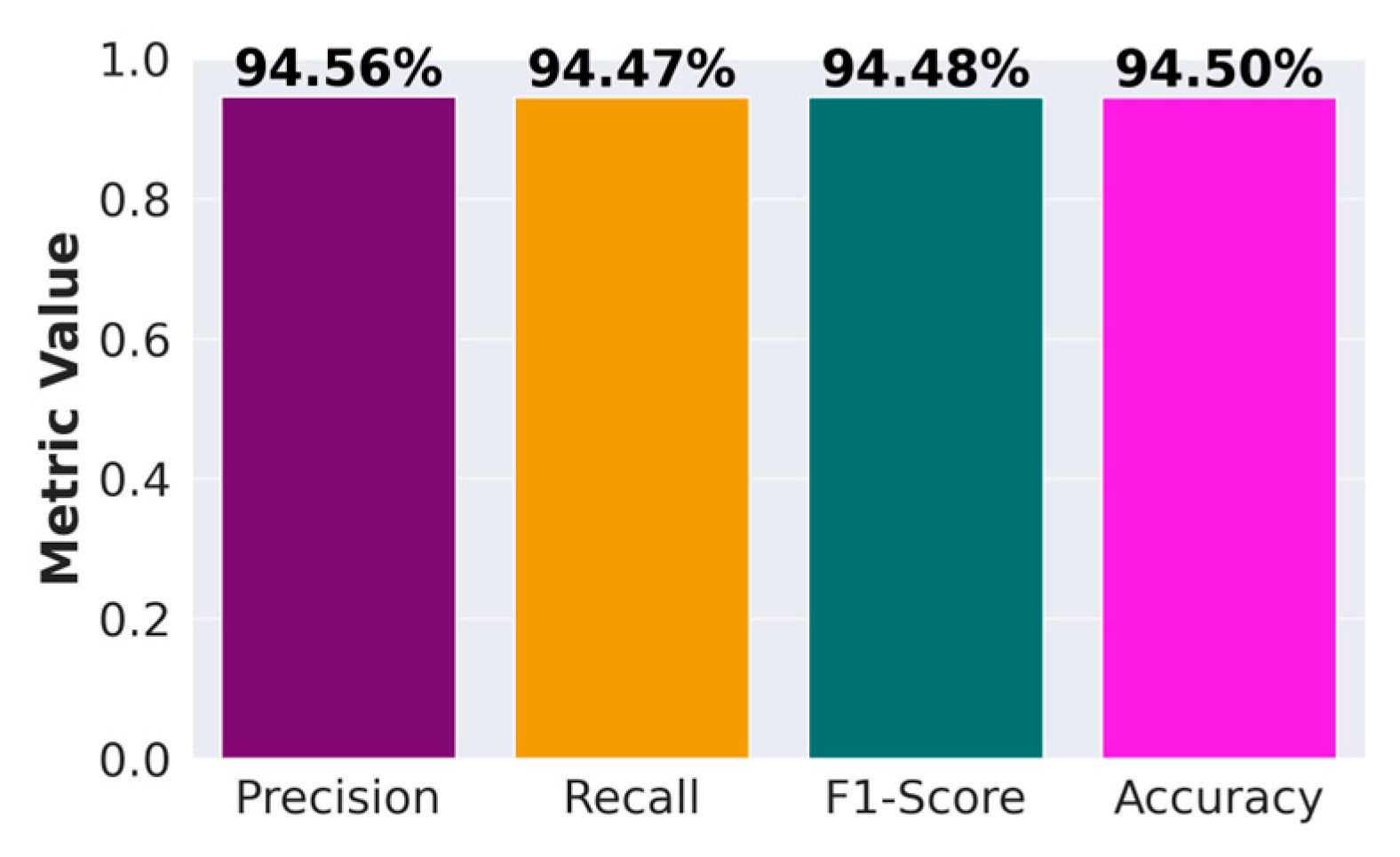
Figure 6 presents a performance comparison of DenseNet, Alexnet, and SqueezeNet. The performance metrics considered are average accuracy, precision, recall and the F1-score.
As we can see from the presented confusion matrices in
Figure 5, very few samples have been misclassified by the DenseNet compared to the baselines.
Figure 6 provides a figure showing the performance of DenseNet against AlexNet and SqueezeNet in terms of accuracy, precision, recall, and F1 score. For all the performance metrics, DenseNet performs better. DenseNet’s architecture allows for each layer to have direct access to the gradients from the loss function and the original input signal, leading to more efficient training and feature reuse. This is particularly useful in histopathological image analysis, where subtle features and patterns are crucial for accurate classification. Furthermore, in DenseNet, each layer receives a “collective knowledge” from all preceding layers, which improves the flow of gradients throughout the network. This leads to better learning and performance, as each layer can learn more complex features based on all previous layers. To further improve the performance, as presented in
Section 3, we adopt ATT-DenseNet, which is our proposed method. We achieved the following performance (
Figure 7), which surpasses the traditional DenseNet, AlexNet and SqueezeNet.
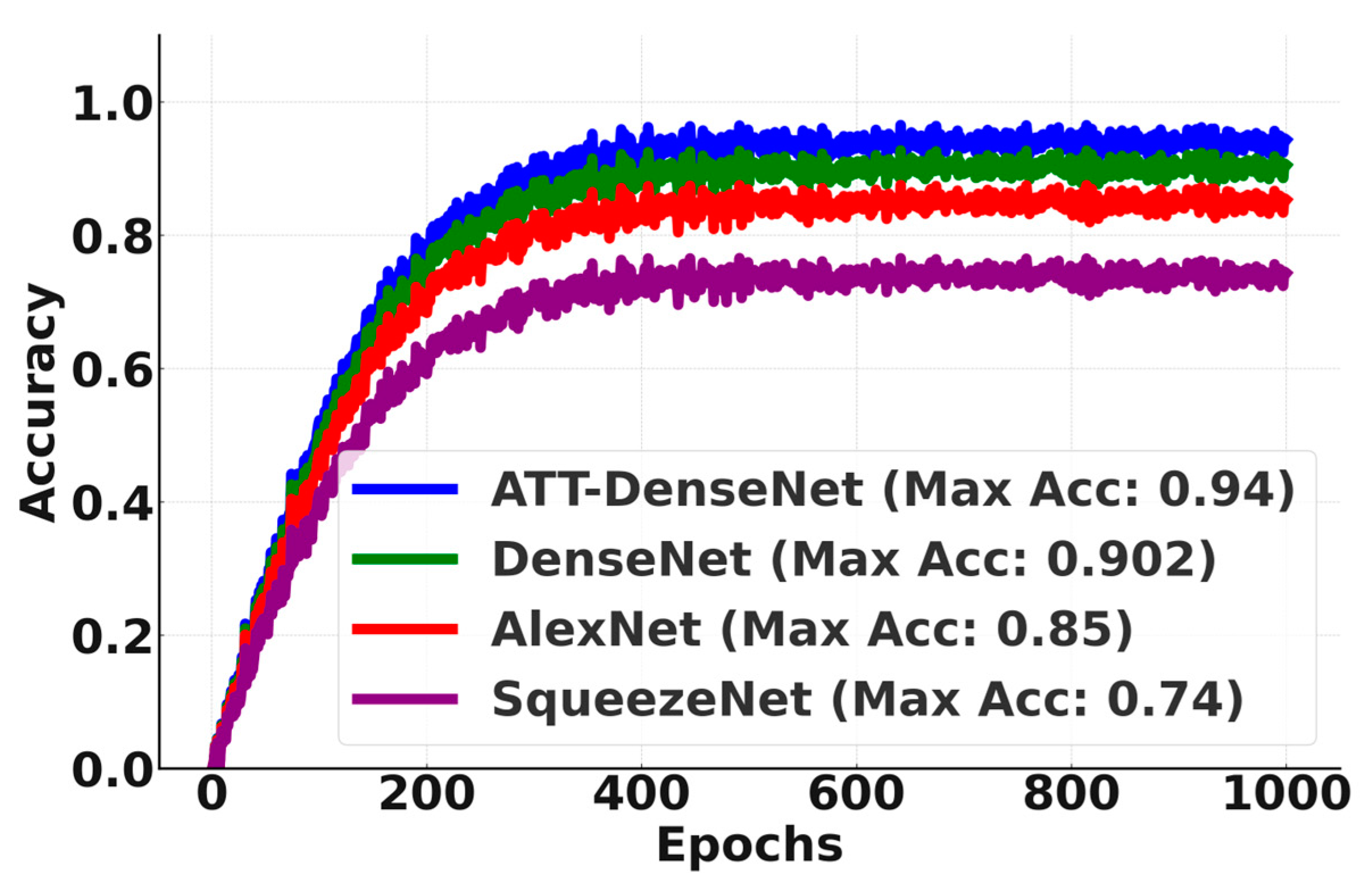
Finally, to show the better performance of ATT-DenseNet in a more vivid way,
Figure 8 presents a comparison of the accuracy curves over 1000 epochs. As we can see from the figure, it outsmarts the baselines by significant margins, achieving an average accuracy of 0.954 (95.4 in percentage).
4.3. CT Scan Images
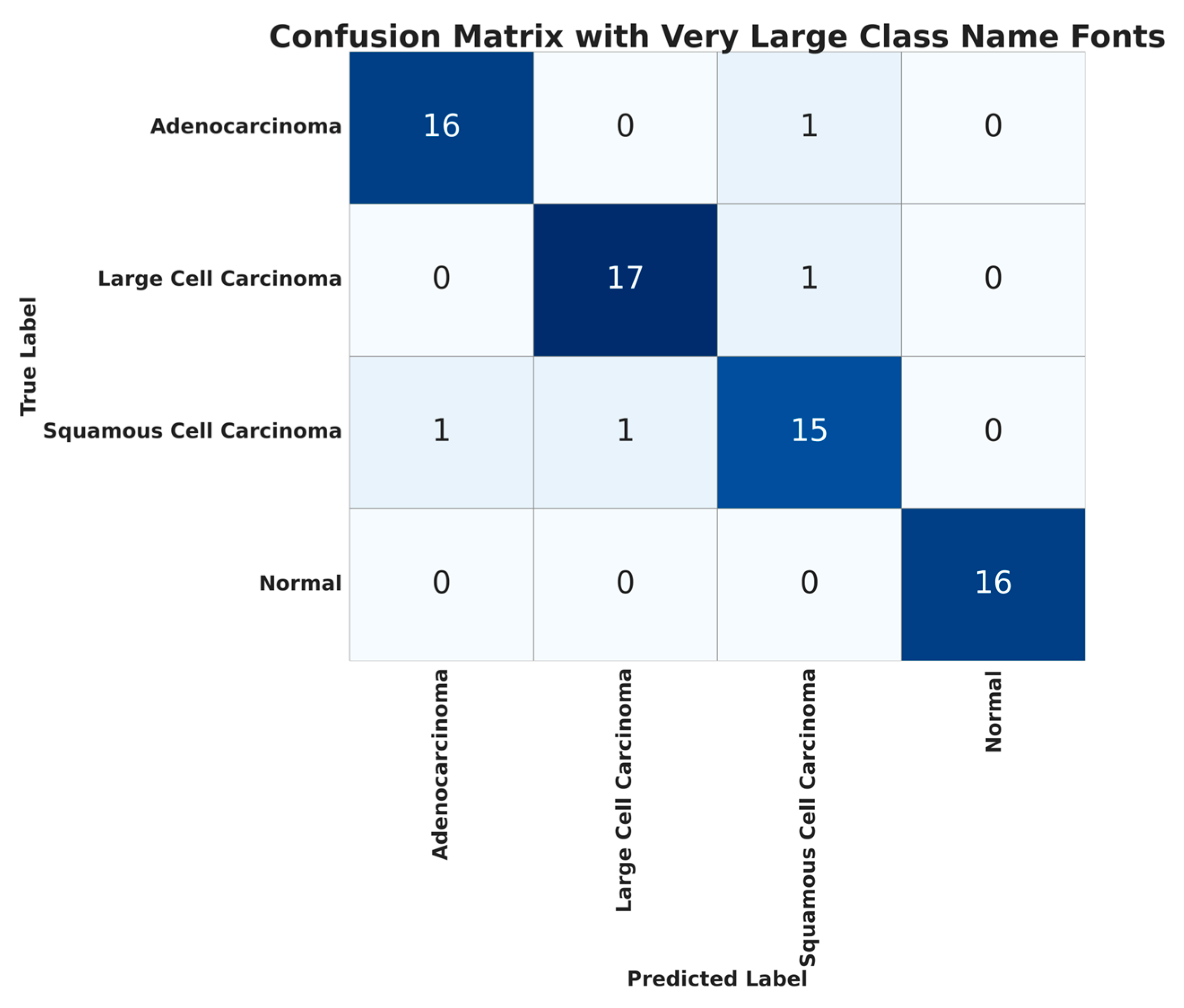
As mentioned in previous sections, the CT scan images used for cancer detection have four classes. The confusion matrix for the classification using ATT-DenseNet is presented in
Figure 9. We do not extensively present the confusion matrices as we did before for histopathological images in order to avoid repetition. Furthermore,
Figure 10 presents an accuracy curve comparison.
We can see from
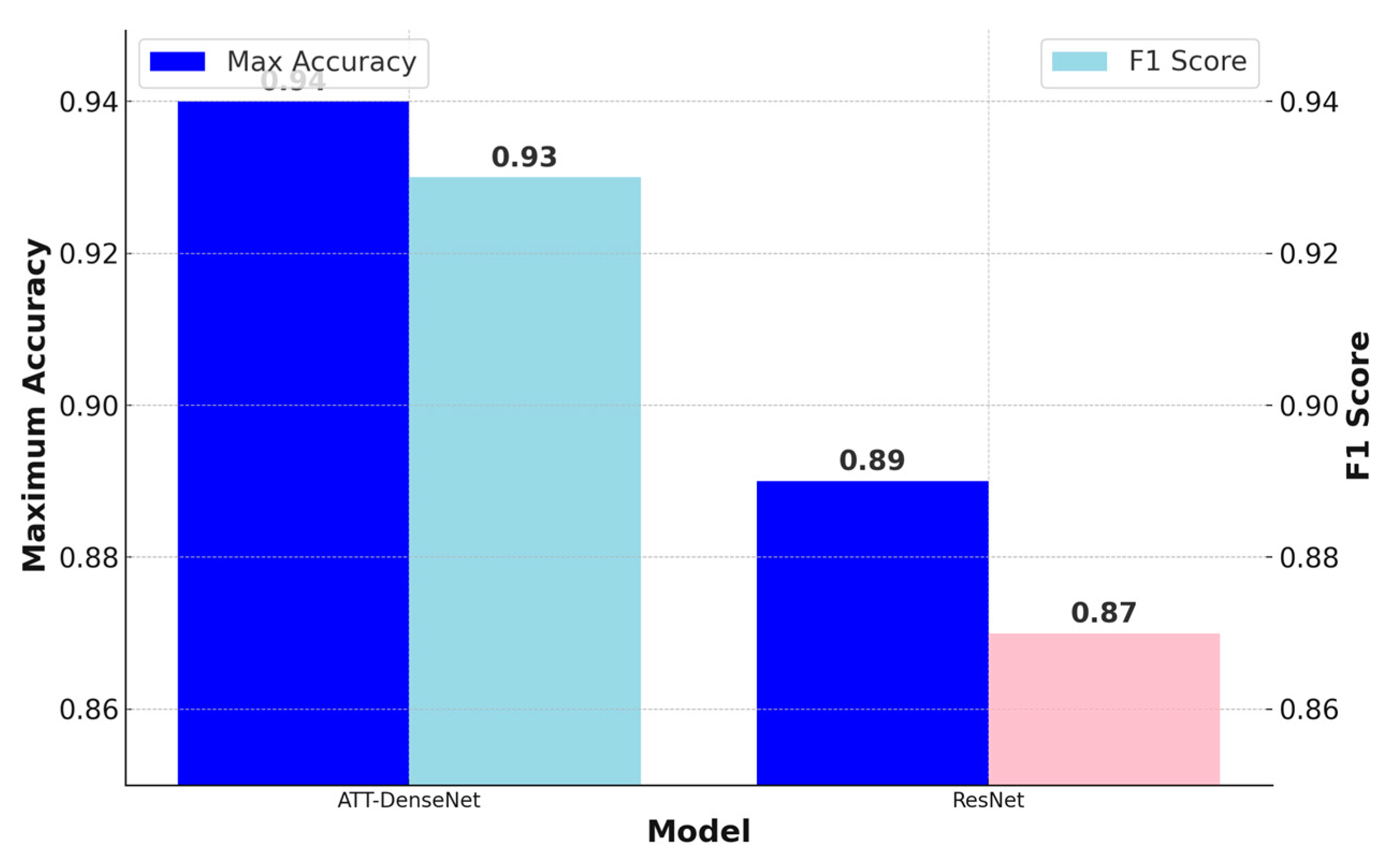
Figure 10 that the proposed ATT-DenseNet achieves higher accuracy compared to all the baselines by achieving 94% average accuracy on the test set of the data. Similarly, as presented in terms of histopathological images, the F1-score of ATT-DenseNet is found to be higher than all the other baselines for CT scan images. Finally, we compare the proposed method with RestNet in terms of accuracy and the F1-score and see that the proposed method performs better as well.
The proposed ATT-DenseNet mechanism adaptively recalibrates feature responses by explicitly modeling interdependencies between channels. This means that the method can dynamically emphasize informative features while suppressing irrelevant ones. By contrast, DenseNet without attention, SqueezeNet, and AlexNet lack such mechanisms to focus on the most relevant features, potentially leading to suboptimal feature utilization. Furthermore, the proposed ATT-DenseNet facilitates better discrimination of features by learning channel-wise relationships. By selectively emphasizing important features, the method can potentially enhance the discriminative power of the network, leading to improved classification performance. DenseNet without attention, SqueezeNet, and AlexNet do not have the capability to learn such discriminative features effectively. The adaptive nature of ATT-DenseNet allows the method to dynamically adjust feature importance based on the input, leading to improved generalization across the datasets and scenarios. This adaptability enables our model to capture complex patterns in the data more effectively compared to the fixed feature mappings of DenseNet without attention, SqueezeNet, and AlexNet. DenseNet’s dense connectivity pattern enables feature reuse throughout the network, leading to parameter efficiency. This means DenseNet requires fewer parameters compared to ResNet or traditional architectures to achieve similar or better performance. With fewer parameters, DenseNet models can be trained faster and require less memory. DenseNet’s dense connections facilitate a direct gradient flow from the later layers to the earlier layers during backpropagation. This helps alleviate the vanishing gradient problem, making it easier to train very deep networks. That is why, as presented in
Figure 8,
Figure 10 and
Figure 11, we achieved better accuracy for the proposed ATT-DenseNet.
4.4. Computational Complexity and Reproducibility
DenseNet typically has a higher computational cost compared to SqueezeNet due to its larger number of parameters, with DenseNet often ranging from 20–30 million parameters, SqueezeNet usually having around 0.7–1.3 million parameters, and AlexNet falling in between, with approximately 60–70 million parameters. This higher parameter count in DenseNet leads to increased memory usage during training and inference, as well as longer training times, especially on datasets with a large number of samples. However, DenseNet’s parameter efficiency and dense connectivity may offer better feature reuse and representation learning, potentially leading to higher accuracy and more robust models, particularly in scenarios with abundant computational resources. This is particularly beneficial in medical applications such as lung cancer classification, where accuracy is paramount and where even marginal improvements can have significant clinical implications. The intricate and nuanced patterns present in medical images require models capable of capturing fine-grained details, which DenseNet’s dense connectivity facilitates. While the computational cost may be higher, the potential for improved accuracy and better performance in medical diagnosis justifies the investment in computational resources. Therefore, despite the higher computational cost, DenseNet is preferred for medical applications where accuracy and reliability are critical, even if it requires more computational resources compared to alternative architectures like SqueezeNet. The results in this section support this statement since ATT-DenseNet, the proposed method, outperforms the other two baselines with higher rates of accuracy.
We tested the reproducibility of the results multiple times with different sets of input extracted from the dataset. In particular, there were 15,000 images in total in the dataset. We took different batches of 750 images to perform the classification to see whether we obtained similar results.
4.5. Clinical Implications and Limitations
The ATT-DenseNet model, designed for lung cancer detection using CT and histopathological images, has significant clinical implications and potential impacts on patient diagnosis and treatment. The model’s innovative approach, which emphasizes relevant image regions and utilizes advanced data preprocessing techniques, sets a new standard in the field. By achieving impressive average accuracies of 95.4% for histopathological images and 94% for CT scan images, ATT-DenseNet not only demonstrates superiority over traditional models like DenseNet, AlexNet, and SqueezeNet but also highlights its potential as a transformative tool in medical diagnostics. This enhanced accuracy and precision in detecting lung cancer could lead to earlier and more accurate diagnoses, enabling timely and personalized treatment plans for patients. Early detection is critical in improving survival rates for lung cancer patients, as it allows for interventions at a stage when the disease is more treatable. Furthermore, the ability of ATT-DenseNet to precisely identify cancerous tissues within images can assist in planning targeted therapies, reducing the need for invasive diagnostic procedures, and ultimately contributing to better patient outcomes. The model’s emphasis on relevant regions within the images ensures that clinicians receive focused and significant diagnostic information, potentially streamlining the decision-making process in clinical settings and improving the efficiency of lung cancer screening programs.
The proposed method showcases significant improvements in detection metrics. However, it faces limitations in generalizability and class imbalance. The model’s performance on diverse datasets is uncertain due to variations in imaging protocols and patient demographics, highlighting the need for models that adapt to different data characteristics. Additionally, the prevalent issue of class imbalance, where non-cancerous images outnumber cancerous ones, can skew the model towards predicting the majority class, potentially reducing its sensitivity to cancerous cases. Addressing these challenges requires further research into robust model design, advanced data augmentation, and balancing techniques to ensure the model’s effectiveness across varied datasets and improved detection of cancerous images. We also want to include some formal methods for AI-based technique verification [
34,
35].
5. Conclusions
In this paper, we successfully developed the ATT-DenseNet model, which notably improved the accuracy, precision, recall, and the F1-score for lung cancer detection using CT and histopathological images. This model’s innovative approach, which emphasizes relevant image regions and utilizes advanced data preprocessing techniques, sets a new standard in the field. The results, with an impressive average accuracy of 95.4% for histopathological images and 94% for CT scan images, not only demonstrate the superiority of ATT-DenseNet over traditional models like DenseNet, AlexNet, and SqueezeNet but also highlight its potential as a transformative tool in medical diagnostics, offering new possibilities for the early and accurate detection of lung cancer. Future research directions could explore further enhancements to the ATT-DenseNet model for even greater rates of accuracy and efficiency in lung cancer detection. Further research could also focus on reducing the model’s computational requirements to facilitate its deployment in resource-limited settings, ensuring wider accessibility and use.