Abstract
Fish possess lipases from embryonic development to adulthood. Lipase activity methods vary and significantly differ in terms of the concentration of the substrate used, bile salt, Ca2+, temperature, pH, and type of lipase units, which limits comparative studies. The three most-used substrates are p-nitrophenyl (p-NP), β-naphthyl (β-N) derivates, and emulsified natural oils. These were selected to be redesigned in this study to measure lipase activity under temperature, pH, ion, and bile salt conditions closer to fish physiology, using the appropriate molar absorption coefficient to calculate the lipase units. Cynoscion parvipinnis (CP), Seriola rivoliana (SR), Centropomus viridis (CV), Elop affinis (EA), and Canthidermis maculate (CM) pyloric caeca-intestine extracts were studied. Sodium taurocholate showed the highest activity for intestinal lipases, and the fatty acid length in the substrates changed the lipase hydrolysis rate. The highest lipase activity was obtained with p-NP butyrate and p-NP caprylate in four fish species. Lipase activity was highly activated with Ca2+ (4–7 mM). The β-N absorption spectrum indicates a plateau between 534 and 554 nm for fish lipases. Salmon oil was identified as the most digestible lipid in the four fish species using the in vitro digestibility assay. The lipase zymogram showed an apparent size of 46.3 kDa for CP, 40.2 kDa for SR, 46.2 kDa for CM, 106.6 kDa for EA, and 58.3, 84.6, and 162.1 kDa for CV.
Key Contribution:
As an alternative approach to optimize fish lipase determination, practical methods at the microplate level are proposed with β-naphthyl caprylate and p-nitrophenyl myristate. In addition, a simple method with olive oil as a substrate is employed in the pH Stat assay.
1. Introduction
Lipases play an important role in activating the different lipids necessary in the different stages of fish development, and in metabolic processes. In this sense, lipids are the primary energy source during larval fish development, growth (fatting), and reproduction; thus, it is paramount to study lipid metabolism. Furthermore, fish lipases constitute an essential molecule from physiological, biochemical, and biotechnological points of view. At present, different methods are used to determine lipase activity. For example, synthetic p-NP derivatives of different chain lengths have been used to assess lipase activity. Among these substrates, p-NP acetate [1,2,3,4,5], butyrate [6], laureate [7], myristate [8], palmitate [9], etc., have been used. However, contrary to long-chain p-NP derivatives, short-chain ones are unsuitable for lipase activity since these substrates are recognized by general esterases, including proteases [10,11]. Fish differ from mammals in their lipolytic enzymes for lipid digestion. In fish, carboxyl ester lipases (CELs) are predominant, which depend on bile salts for activity and have the broadest substrate specificity, in contrast to pancreatic lipases (PLs) in mammals, which depend on bile salts and colipase [12]. Marine fish, like the ones tested in this work, predominantly possess CELs, compared to freshwater fish, which predominantly possess PL-like enzymes. Since marine fish are primarily piscivorous, they obtain their energy from various lipid classes. In contrast, many freshwater fish species are principally omnivorous, so they obtain energy from lipids and carbohydrates [13].
The effect of detergents is not homogeneous for all lipases [14]. In addition, the influence of these compounds on lipase activity is dose-dependent [15]. Bile salts are key activators for mammal pancreatic lipases in the presence of colipase [16,17] and are inhibitors for other lipases [18]. SDS, known for its denaturant effect on proteins, activates several lipases and inhibits others [19]. Some non-ionic detergents avoid the formation of enzymatic aggregates [20] and structurally stabilize the molecule [21]. Triton-X100 is a non-ionic detergent that some authors have reported as a stimulator of lipase activity [22], but it can also inhibit fish lipases [23]. In this work, we propose avoiding the use of artificial detergents and use natural bile salts instead.
Methods to measure lipase activity are still being discussed because of the different conditions required. Nolasco-Soria [24] found that the methods used by authors are very varied and have significant differences in terms of the type of substrate, substrate concentration, bile salt type and concentration, calcium concentration, pH, temperature, incubation time, measurement of hydrolysis products, and definition of lipase units. The present research intends to optimize current methods to measure the activity of lipases (glycerol-ester hydrolases; EC 3.1.1.3) that catalyze the hydrolysis of ester bonds in acylglycerols in vivo [25]. These enzymes could be selective for lipid type [26] and the position and type of fatty acids, including chain length [27,28,29]. These enzymes could also be stereoselective [30], differentiating between enantiomers, towards racemic substrates, or between enantiotropic groups for prochiral triacyl glycerides [31]. Finally, certain combinations of previous selectivity types could be established [32]. Synthetic acyl chain esters of p-nitrophenol and α/β-naphthol substrates have been used to assess lipase activity for decades, following the release of the alcoholic base by spectrophotometric methods [33]. However, short-chain p-nitrophenyl (p-NP) or β-naphthyl (β-N) derivatives, such as p-NP acetate, should be avoided due to their non-specific hydrolysis by general esterases, rather than by true lipases [1,11].
The most-used method for measuring lipase activity in fish is p-nitrophenyl (p-NP) substrates, mainly p-NP myristate. According to Nolasco-Soria [24], the method most used is the one published by Iijima et al. [34]. p-NP myristate is selected to optimize lipase activity measurements. After verifying the main methodological variants (calcium concentration and type and concentration of bile salts), it is considered necessary to experimentally evaluate the environmental and composition variants of the reaction mixture to support the proposal of a method as an alternative to standardization. The method must be safe, sensitive, and practical at the microplate level, considering the presence of bile salts and calcium at the appropriate concentrations. Because of the lack of studies about optimum calcium and bile salt concentrations in the fish digestive tract, the appropriate concentrations will be considered to give maximum activity in the assay of the tested fish used as an example in this study. The use of a wavelength provides greater sensitivity, and the molar absorption coefficient (ε) is used according to the corresponding standard curve.
According to Nolasco-Soria [24], the third most-used method for fish lipase activity assays is the one published by Versaw et al. [35], using β-N caprylate as a substrate. In general, the methods for lipase activity measurement using β-naphthyl substrates are analyzed to make optimization proposals towards standardization, considering β-naphthol absorption spectra, standard curve, and the effect of pH on color development.
Nolasco-Soria [24] and Bier [36] published the second most-used method for measuring lipase activity, using olive oil as a substrate. The measurement of lipase activity using natural oils as a substrate in a small volume (5 mL) is proposed as a definitive test to confirm the presence of lipases in the digestive tract of fish. Lipase activity can be assessed by following the release of fatty acids from a natural substrate (triacyl glyceride of long-chain fatty acids). This technique can be performed indirectly by observing the released protons during the enzymatic hydrolysis. The pH Stat method, based on the titration of pH decrease in the function of time, is one of the most-used techniques for this principle [37].
Metallic ions can affect lipase activity in different manners, and can stabilize (or activate) (e.g., zinc on a thermostable Bacillus stearothermophilus lipase) [38] or destabilize (or inhibit) (e.g., mercury on a Channa punctatus lipase) [39] the structure of these enzymes in solution. Many lipases require Ca2+ to maintain a stable and/or catalytically competent conformation [28,40]. In addition, metallic cations could also act as activators of lipases [16].
Detergents or surfactants, such as bile salts, are additives of great importance for lipolytic activity [9], since they are involved in the formation of micelles from the dispersion of large hydrophobic aggregates on which the natural substrates of these enzymes are spontaneously organized in aqueous solution. This organization causes a notable increase in the superficial area available for enzyme–substrate interaction, with a resultant effect on reaction velocity [41]. Detergents could also influence the stereoselective properties of lipases [42]. Due to the variability in the methods used to measure lipase activity in aquatic organisms [24], and to standardize measurement protocols performed outside the physiological conditions of the fish, we propose a new method to measure lipase activity in samples from the digestive tract of fish found on the Baja California Sur coast that are essential in local fisheries, such as the shortfin weakfish (Cynoscion parvipinnis, CP), longfin yellowtail (Seriola rivoliana, SR), white snook (Centropomus viridis, CV), machete (Elops affinis, EA), and rough triggerfish (Canthidermis maculate, CM). The method involves a pH closer to the intestinal pH of fish (8.0) [43] and is performed at 25 °C, closer to the ecophysiological temperature of temperate water for fish [44]. We also determined the optimum calcium and bile salt concentrations for optimal activity and present a native electrophoretic method for identifying proteins with lipase activity.
2. Materials and Methods
2.1. Reagents and Materials
The reagents and materials obtained from Sigma-Aldrich (St. Louis, MO, USA) included the following: sodium cholate (C1254), sodium choleate (S9875), Trizma (T6066), dimethyl sulfoxide (D8418), Fast Blue BB Salt hemi (zinc chloride) salt (F3378), trichloroacetic acid (T6399), sodium dodecyl sulfate (L6026), β-naphthol (185507), 4-nitrophenyl myristate (70124), 4-nitrophenyl butyrate, 4-nitrophenyl caprylate, 4-nitrophenyl palmitate, 4-nitrophenol (1048), β-naphthyl caprylate (β-NC) (N-100-1), sodium hydroxide (S5881), sodium taurocholate (S-121-50) (from Gold Biotechnology, St Louis MO, USA), a Bio-Rad Protein Assay Kit II (5000002, from Bio-Rad, MX, Hercules, CA, USA), and Grainer 96-well plates (M2936) (Greiner products from Merck KGaA, Darmstadt, Germany, DE). Olive oil (City Club Members choice, extra virgin), krill oil (Kirkland), and salmon oil (Nodrim) (food-grade) from commercial brands were obtained from local commercial stores in La Paz, BCS, Mexico.
2.2. Fish Sample Testing
To evaluate the performance of the lipase procedures, pyloric caeca-intestine or empty intestine samples (from pools of 3 fishes) were obtained from shortfin weakfish (Cynoscion parvipinnis, CP), longfin yellowtail (Seriola rivoliana, SR), white snook (Centropomus viridis, CV), machete (Elops affinis, EA), and rough triggerfish (Canthidermis maculata, CM), and treated as per Nolasco-Soria’s description [45]. All fish were obtained from local fishermen; immediately after fishing, they were sacrificed by an overdose of MS-222 (tricaine metasulfonate) and kept on ice until landing, where they were immediately transferred to the laboratory (NOM-062-ZOO-1999, 2001). Tissues were used for the preparation of lipase crude extract (CE). The extraction procedure in all cases was as follows: The tissue was placed in a beaker within an ice bath (0–4 °C); then, 3 volumes (w:v) of 0.025 M universal buffer, pH = 8.0 (4 °C), was added, and the mixture was homogenized for 60 s three times (the use of a blade homogenizer like ULTRA-TURRAX© at 24,000 rpm is recommended). The homogenates were clarified by centrifugation at 15,000× g for 15 min at 4 °C. Subsequently, they were centrifuged again at 15,000× g (15 min at 4 °C), and the supernatants were stored in 2 mL aliquots at −80 °C until their further analysis for lipase activity. The CE protein concentration was quantified using a bovine albumin standard curve [46]. The fish digestive tract sections’ somatic indices and protein concentrations (Table 1) were calculated by dividing each tissue’s fresh weight by the fish’s fresh weight (precision 0.0001 g).

Table 1.
Fish weight, somatic index, and extract protein concentration.
2.3. Test towards the Standardization of Lipase Methods
As a starting point for standardization, the lipase protocols reported by Nolasco-Soria et al. [23] (for the substrates p-NP butyrate, p-NPB; p-NP caprylate, p-NPC; p-NP myristate, p-NPM; p-NP palmitate, p-NPP; β-N caprylate, β-NC; and natural oils) were taken as a base, mainly to determine the effects of calcium and bile salt concentrations on lipase activity measured by spectrophotometric methods. In line with Nolasco-Soria [45,47], for fish alkaline protease activity and fish amylase activity, the pH and reaction temperature in the fish digestive enzyme methods were adjusted to pH 8.0 and 25 °C, respectively. All lipase determinations included a blank reference test, in which the CE was replaced by distilled water, or was inactivated by heat treatment at 100 °C for 10 min. In the spectrophotometric methods, the absorbance units of the blank reference tests were always subtracted from those produced by the corresponding experimental active lipase extracts. All absorbance values obtained in the microplate reader, using 96-well microplates, were adjusted to a light length of 1 cm. All experimental assays were completed at least in triplicate.
2.4. p-NP Substrate Method
Enzymatic assay: According to Nolasco et al. [23], lipase activity was kinetically determined using p-NP substrates. In a 96-well microplate, each assay (200 µL) contained 0.5 mM p-NP substrate, 10 mg mL−1 (18.6 mM) sodium taurocholate (NaT), and 20 mM Tris-HCl (pH 8.0). Typically, 10 μL of CE was added to the reaction mixture. After 10 s of shaking (at 300 rpm, 15 mm diameter), incubation was carried out for 10 min at 25 °C, and the absorbance was read at 400 nm (according to the absorption spectrum of the reaction mixture from the p-NP standard curve) every 30 s (Varioskan Flash, Thermo Scientific, Waltham, MA, USA). One unit of lipase activity was defined as the enzyme amount that generated 1 μmol of p-NP per minute.
2.5. p-NP Absorption Spectrum
Standard solutions of p-NP in DMSO were prepared. A 200 µL solution of 0.1 mM p-NP in DMSO with an absorbance close to 1.0 at 400 nm was selected to make its absorption spectrum of pure p-NP between 300 and 600 nm. The reaction mixture included 10 μL of substrate (diluted in DMSO) in a final volume of 200 μL. Then, the final concentration of DMSO (by substrate) was 5% v/v. The absorption spectra of the actual final reaction mixtures in the presence of an enzyme (Seriola rivoliana intestine extract) and the blank (without enzyme) were also determined.
2.6. p-NP Standard Curve
The p-NP standard curve was also constructed in a total volume of 200 µL, using 10 mg mL−1 (18.6 mM) NaT, 20 mM Tris-HCl (pH 8.0), 4 mM calcium chloride, and 0.1 mM NaCl, adjusting the concentration of p-NP from 0 to 0.4 mM in the reaction mixture.
2.7. Effect of Type of Bile Salt
The effect of the type of bile salt was determined under the same experimental conditions using sodium cholate, choleate [48], or taurocholate (10 mg mL−1 final concentration) [49].
2.8. Effect of Sodium Taurocholate (NaT) Concentration
The effect of NaT concentration on lipase activity was evaluated under the same experimental conditions, but the NaT concentration was adjusted from 0 to 20 mg mL−1, and p-NP butyrate (4 C), caprylate (8 C), myristate (14 C), and palmitate were used (16 C).
2.9. Effect of CaCl2 Concentration
The effect of CaCl2 concentration on lipase activity was evaluated under the same experimental conditions, but including CaCl2 at concentrations from 0 to 10 mM.
2.10. β-NC Method
Enzymatic assay: Lipase activity, using β-NC as the substrate, was endpoint-determined spectrophotometrically, according to Nolasco et al. [23]. In a 96-well microplate, each assay (100 µL) contained 1 mM β-NC, 3 mg mL−1 (5.6 mM) NaT, 20 mM Tris-HCl (pH 8.0), 1 mM Fast Blue BB salt. Typically, 10 μL of CE was added to the reaction mixture. After 10 min incubation at 25 °C, the reaction was stopped by an SDS-TCA reagent (100 µL), and the absorbance was read at 540 nm (according to the absorption spectrum of the reaction mixture from the β-N standard curve) (Varioskan Flash, Thermo Scientific). One unit of lipase activity (vs. β-NC) was defined as the enzyme amount that generated 1 μmol of β-N released per minute.
2.11. β-N Standard Curve
A β-N standard curve was prepared under the same experimental conditions of the indicated reaction mixture, substituting the CE for β-N at a final concentration of 0 to 0.1 mM and adding 100 mM NaCl and 4 mM CaCl2. The absorption spectra of the actual final reaction mixtures in the presence of an enzyme (Seriola rivoliana intestine extract) and the blank (without enzyme) were determined. The standard curve was constructed, replacing the Tris-HCl buffer with universal buffer (50 mM) at pH 6, 7, 7.5, 8, 9, and 10 to demonstrate the effect of pH on the β-N-Fast Blue BB colored complex.
2.12. Natural Oil pH Stat Method
To fulfill the requirement of using natural lipids as a substrate to declare a true lipase, as established by Cherry and Crandall [50], the proposed method to measure lipase activity includes using olive oil as a substrate and evaluating the release of protons with the pH Stat method [23,51]. The lipase activity was determined by the micromoles of NaOH required per minute to maintain the pH at 8.0 in the reaction mixture (which had an initial volume of 5 mL) during 15 min of hydrolysis at 25 °C. One lipase unit was the enzyme amount required to release one micromole of fatty acid per minute.
2.13. Oil In Vitro Digestibility
The in vitro digestibility of alimentary oils (vegetable, crustacean, and fish) was determined by using the CE of the studied fish as the enzymatic reagent, following the previously described method. In the in vitro digestibility tests of natural oils, it was verified that during the established digestion time, the stability of the emulsions was not altered, as there was no apparent separation of the water–oil phases. All the in vitro hydrolysis (digestion) tests included a control group where the CE was previously inactivated by heat treatment (water bath at 95 °C for 10 min) and a control treatment where the CE was substituted for distilled water. A thermostat jacket was used to keep the assay temperature at 25 °C. All the analyses were determined in triplicate.
2.14. Native Lipase Electrophoretic Analysis
The development of lipase isoenzymes in electrophoresis gels (running at native conditions) was carried out as follows: Gradient polyacrylamide gels (4–20%) were prepared (a 4–15% polyacrylamide gradient gel can be used for a significant displacement of proteins in the gel, particularly for high-molecular-weight protein clusters). The gels were placed in the electrophoresis chamber (Mini-PROTEAN Vertical Electrophoresis Cell, BIORAD) and equilibrated with a running buffer (Tris-Glycine, pH 8.3) for 15 min at 80 volts. Extracted volumes of CP, SR, CM, EA, and CV equivalent to 100 µg of protein were applied with bromophenol blue (0.1%) and sucrose (10%). The gels were run at 120 V until the bromophenol blue marker reached the edge of the gel. A molecular weight marker was also used (Invitrogen™ NativeMark™ Unstained Protein Standard, LC0725, Thermo Fisher Scientific, USA). The gel was disassembled from the chamber and incubated in a substrate solution (19 mL of 20 mM Tris-HCl, pH 8, containing NaT at a final concentration of 10 mg mL−1 (18.6 mM) + 1 mL of 10 mM β-NC substrate, in DMSO (reaction concentration of DMSO was 5%)). The gel was kept inside a black plastic bag to reduce dehydration. Then, the gel bag was placed in the cold (refrigerator at 4 °C) for 5 min; then, it was placed in a water bath for 10 min at 35 °C. The gel was removed and covered with the developer solution (19 mL of 20 mM Tris-HCl, pH 8, containing NaT at a final concentration of 10 mg mL−1 (18.6 mM) + 1 mL of 10 mM Fast Blue BB (FBBB), in DMSO). The formation of red bands was monitored and photo-documented by photography on a white illuminated surface or photo-documentation (BIORAD, Gel Doc XR+). Subsequently, the gel was stained for protein with Coomassie Blue solution (0.1%) in methanol–acetic acid–water (40:10:50 v:v:v) for 45 min, destained with methanol–acetic acid–water overnight, and photo-documented as indicated.
2.15. Statistical Analysis
The results are reported as means ± SEM (standard error of the mean). Data were checked for normality and variance homogeneity and were evaluated using the ANOVA test, followed by the post hoc Tukey–Kramer multiple comparisons test. The level of significance was set at p < 0.05.
3. Results
3.1. Absorption Spectrum and Standard Curve with p-NP
The p-NP absorption spectrum indicates the absorption peak at 400 nm, both for the pure p-NP and the actual reaction mixture (Supplementary Figure S1). The p-NP standard curve at 400 nm gives an equation of y = 16756x (R2 = 0.999) (Supplementary Figure S2).
3.2. Effect of Type of Bile Salt on Lipase Activity in p-NP Substrates
Figure 1 shows the effect of the type of bile salt on lipase activity when using p-NPM as a substrate. In the four fish used as experimental models, sodium taurocholate (NaT) presented the highest activity for intestinal lipases. Different letters indicate statistical differences in all figures.
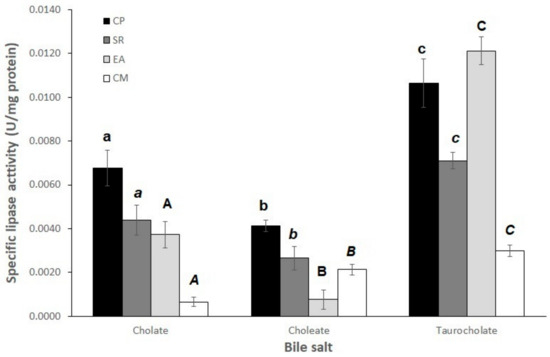
Figure 1.
Effect of bile salts on intestinal lipase activity in shortfin weakfish (Cynoscion parvipinnis, CP), longfin yellowtail (Seriola rivoliana, SR), machete (Elops affinis, EA), and rough triggerfish (Canthidermis maculata, CM). For each fish species, different letters for each fish and different bile salts indicate significant differences (p < 0.05).
3.3. Effect of NaT Concentration on Lipase Activity in p-NP Substrates
The effect of NaT concentration on lipase activity is shown in Figure 2. In general, lipase activity in p-NP substrates with fatty acids of short or medium length (4C and 8C) was not activated by NaT. For the 4C substrate, it was inhibitory. For the 8C substrate, it had no significant effect. In contrast, for long substrates (14C or 16C), it had an activating effect at concentrations of NaT until 20 mg/mL in all CE species. It should be considered that the length of the fatty acid in the substrates of the p-NP series changes the rate of hydrolysis of the lipases. Figure 3 shows the specific activity of the extracts from five fish species using the substrates p-NPB, p-NPC, p-NPM, and p-NPP, with 10 mg mL−1 (18.6 mM) NaT. The highest activity was obtained with p-NPB and p-NPC, except for the CM extract, for which the most increased activity was observed with p-NPM.

Figure 2.
Effect of NaT concentration on lipase activity in shortfin weakfish (Cynoscion parvipinnis, CP), longfin yellowtail (Seriola rivoliana, SR), white snook (Centropomus viridis, CV), machete (Elops affinis, EA), and rough triggerfish (Canthidermis maculata, CM). Different letters show significant differences (p < 0.05) between temperatures and NaT concentration.

Figure 3.
Lipase activities of the extracts from shortfin weakfish (Cynoscion parvipinnis, CP), longfin yellowtail (Seriola rivoliana, SR), machete (Elops affinis, EA), and rough triggerfish (Canthidermis maculata, CM) using the substrates p-NP butyrate, p-NP caprylate, p-NP myristate, and p-NP palmitate. Different letters show significant differences (p < 0.05) between species and fatty acid length.
3.4. Effect of CaCl2 Concentration on Lipase Activity in p-NP Substrates
Figure 4 shows the effect of Ca2+ concentration on lipase activity. Lipase activity in the p-NPM substrate was generally activated at a Ca2+ concentration between 4 and 7 mM.

Figure 4.
Effect of Ca2+ concentration on lipase activity in shortfin weakfish (Cynoscion parvipinnis, CP), longfin yellowtail (Seriola rivoliana, SR), machete (Elops affinis, EA), and rough triggerfish (Canthidermis maculata, CM). Different letters show significant differences (p < 0.05) between species and CaCl2 concentration.
3.5. Absorption Spectrum and Standard Curve with β-N
The β-N absorption spectrum indicates the absorption peak at 540 nm, both for the pure β-N and the actual reaction mixture, with a broad plateau between 534 nm and 554 nm (Supplementary Figure S3). The β-N standard curve at pH 8 (using Tris buffer and universal buffer) at 540 nm gives an equation of y = 24,286x for Tris buffer and y = 24,348x for universal buffer (Supplementary Figure S4). The effect of pH (using a universal buffer) on the absorbance of the β-N-Fast Blue BB colored complex considering the function of concentration is shown in Supplementary Figure S5. The slope of the standard curves increases with the function of pH.
3.6. Lipase Activity Using Olive Oil as Substrate—pH Stat Method
The specific lipase activities of the fish extracts (U/mg protein) using olive oil emulsion as the substrate are shown in Figure 5 for CP (0.0506 ± 0.0078), SR (0.0661 ± 0.0054), EA (0.0797 ± 0.0164), and CM (0.0230 ± 0.0077) extracts. The in vitro oil digestibility for the four fish species is shown in Figure 6. In all cases, salmon oil turned out to be the most digestible.
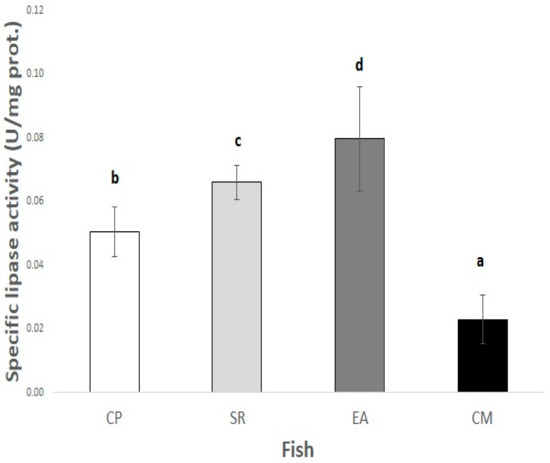
Figure 5.
Lipase activity from shortfin weakfish (Cynoscion parvipinnis, CP), longfin yellowtail (Seriola rivoliana, SR), machete (Elops affinis, EA), and rough triggerfish (Canthidermis maculata, CM) extracts using olive oil emulsion. Different letters show significant differences (p < 0.05) between species.
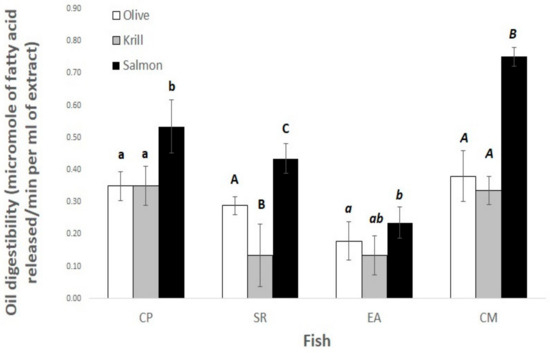
Figure 6.
Lipase activity in shortfin weakfish (Cynoscion parvipinnis, CP), longfin yellowtail (Seriola rivoliana, SR), machete (Elops affinis, EA), and rough triggerfish (Canthidermis maculata, CM) extracts using different oil emulsions. Different letters show significant differences (p < 0.05) between species and oils.
3.7. Native Lipase Isoforms
The gels showing the native isoforms with lipase activity (in the presence of bile salt) from the fish extracts are shown in Figure 7. Under the established native conditions, the apparent sizes (estimated with the obtained molecular-weight standard curve equation of y = −1.9164x + 5.6939, R2 = 0.9808) for the lipase activity bands are about 46.3 kDa for CP, 40.2 kDa for SR, 46.2 kDa for CM, 106.6 kDa for EA, and 58.3, 84.6, and 162.1 kDa for CV.

Figure 7.
Lipase zymograms from (1) CP, Cynoscion parvipinnis; (2) SR, Seriola rivoliana; (3) CM, Canthidermis maculate; (4) EA, Elops affinis; and (5) CV, Centropomus viridis. (A) Photographic image of the electrophoresis gel (PAGE gradient 4–20%), indicating the bands with lipase activity in pink-red color (zymography). (B) Photographic image of the gel stained for protein with Coomassie Blue. The arrows indicate the proposed position of the protein band with lipase activity. MWM: molecular weight markers (used as a reference for the apparent size of protein formations with lipase activity).
3.8. Proposed Final Protocols for Standardization of Lipase Activity Measurement Using p-NPM, β-NC, and Natural Oil as Substrates
3.8.1. p-NPM Method
Lipase activity can be kinetically determined with a spectrophotometer, using p-NPM as a substrate. In a 96-well microplate, each assay (200 µL total volume; five replicates) should contain the following reagents, added in this order: 50 µL of 40 mg mL−1 (74.4 mM) NaT, 120 µL of 50 mM Tris-HCl (pH 8.0), 10 µL of 80 mM CaCl2, and 10 µL of enzyme extract. The mixture should then be stirred on the table in a figure-eight (8) pattern. To start the reaction, add 10 µL of 10 mM p-NPM substrate, and after 10 s of shaking (at 300 rpm, 15 mm diameter), carry out incubation for 5 min at 25 °C. The absorbance at 400 nm should be read every 30 s (Varioskan Flash, TermoScientific). One unit of lipase activity is defined as the enzyme amount that generates 1 μmol of p-NP per minute. The calculation method is as follows: (a) The slope of each reaction mixture (RM) (five samples and three controls) is calculated in absorbance units at 400 nm min−1. (b) The mean slope of the controls is subtracted from the slope of the samples. (c) The absorbance/min value is adjusted to a 1 cm light path (the volume of the RM is 200 μL and gives a liquid column of less than 1 cm). (d) The micromolar concentration of p-NP from the RM is calculated by dividing the absorbance min−1 by the ε (the standard curve to calculate the ε must be constructed at the same working pH used for lipase activity). (e) The μmol of p-NP min−1 (lipase units) is calculated in the RM (0.0002 L). (f) Lipase units mL−1 in the enzyme extract are calculated. If this value is divided by the protein concentration of the enzyme extract, the lipase-specific activity (U mg of protein−1) is obtained. U fish−1 = U mL−1 × total theoretical volume of fish intestine extract; U g of fish−1 = total U of fish g of fish−1; U g of intestine−1 = total U of tissue extract g of the intestine−1 (fresh weight).
3.8.2. β-NC Method
Lipase activity, using β-NC as a substrate, can be endpoint-determined spectrophotometrically. In a 96-well microplate, each assay (200 µL total volume) (five replicates) should contain the following reagents, added in this order: 20 µL of 15 mg mL−1 (27.9 mM) NaT, 140 µL of 50 mM Tris-HCl (pH 8.0), 10 µL of 1 M NaCl, 10 µL of 40 mM CaCl2, and 10 µL of enzyme extract. Then, shake with a movement in the shape of an 8 on the laboratory bench. Add 10 µL of 10 mM β-NC substrate to start the reaction. After 9 min incubation at 25 °C, add 10 µL of Fast Blue BB color developer, followed by an additional 1 min incubation, to obtain a 10 min total incubation. After 10 s of shaking (at 300 rpm, 15 mm diameter), the absorbance at 540 nm should be read (Varioskan Flash, TermoScientific). One unit of lipase activity is defined as the enzyme amount that generates 1 μmol of β-N released per minute. The calculation method is as follows: (a) The absorbance at 540 nm of each RM (five samples and three controls) is obtained. (b) The absorbance of the controls (average) is subtracted from the absorbance of the samples. (c) The absorbance/min is calculated. (d) The absorbance value is adjusted to a 1 cm light path. (e) The micromolar concentration of β-N from the RM is calculated (dividing the absorbance/min by the ε. (f) The micromoles of β-N/min (lipase units) are calculated in the RM (0.0002 L). (g) Lipase U mL−1 of enzyme extract is calculated. Specific activity, U fish−1, U g of fish−1, and U g of the intestine−1 are calculated as previously indicated.
3.8.3. Olive Oil with pH Stat Method
Lipase activity can be kinetically determined using olive oil as a substrate using the pH Stat method. In a 10 mL beaker, each assay (5 mL total volume) (four replicates) should contain the following reagents, added in this order: (a) 1 mL of 62.5 mg mL−1 (116.2 mM) NaT; (b) 3.0 mL of olive oil emulsion (2.5 g of Arabic gum, 48 mL of distilled water, and 2 g olive oil, prepared by Ultraturrax treatment—Ultraturrax PRO 200, 12,000 rpm, 5 min—and sonication treatment—Sonicator Qsonica VWR, 40% amplitude, 60 s × 3 times); (c) 0.5 mL of 0.5 M NaCl; (d) 0.4 mL of 50 mM CaCl2; and (e) 0.1 mL of distilled water. After mixing, the pH is adjusted to 8.0. To start the reaction, add 0.1 mL of enzyme extract and measure the consumption of NaOH (10 mM) by the pH Stat equipment to maintain the pH at 8.0 for 15 min of hydrolysis at 25 °C at 150 rpm (magnetic rod). One lipase unit is the enzyme amount that generates one μmol of fatty acid/min. The calculation method is as follows: (a) The μmol of NaOH consumed per min of each RM−1 (four samples and two controls) is calculated. (b) The μmol of NaOH min of the controls−1 (average) is subtracted from the NaOH min consumption of the samples−1. (c) The μmol of free fatty acids min−1 (lipase units) in the RM (0.005 L) is obtained. (d) Lipase units per mL of enzyme extract−1 are calculated. Specific activity, U fish−1, U g of fish−1, and U g of the intestine−1 are calculated as previously indicated.
A comparison between the classical lipase activity methods and the optimized lipase activity methods is presented in Table 2.

Table 2.
Comparing classical and optimized lipase activity methods.
4. Discussion
The type of bile salt could influence lipase activity because of its well-known role in lipid digestion and absorption. The highest specific lipase activity towards taurocholate, regarding cholate or choleate, in four fish species (Figure 1) is consistent with the spiny dogfish pancreatic lipase, with an activity increased several-fold by sodium taurocholate, but not by deoxycholate [52]. In addition, the CELs from red sea bream, Pagrus major, are activated by taurocholate and choleate, but not by deoxycholate [34]. According to Løkka et al. [53], the in vivo bile salts in salmonid intestinal chyme range between 1 and 8 mg/mL. Bile salts are present at high levels in the chyme of the fish intestine (e.g., salmon and rainbow trout), primarily as taurocholate (>90%). We tested concentrations between 0 and 20 mg/mL (Figure 2) above the indicated bile salt concentration. According to Figure 2, 10 mg/mL (18.6 mM NaT) is the minimum concentration to achieve maximum lipase activity. It has been demonstrated that bile salt activates digestive enzyme activity; intestinal trypsin and lipase activities were significantly increased by the 0.3% inclusion of bile salt in feed to promote feed digestion in juvenile leopard coral grouper (Plectropomus leopardus), according to Gao et al. [54].
Taurocholate is the most extended bile salt among the three tested in this work (Supplementary Figure S6). This feature probably favors the formation of a mixture of micelles with p-NPM substrate, activating lipase activity to a higher extent (Figure 1).
The activation of fish lipases by NaT in long-chain p-NP substrates (14 and 16C, Figure 2) is consistent with the requirement of CELs for bile salts to hydrolyze bulk triacyl glycerides [55]. This activating effect was observed in the range of 10–20 mg/mL NaT (~20–40 mM) (Figure 2). This agrees with a lipase isolated from rainbow trout, which is activated by bile salts in the range of 25–250 mM for triolein (contains fatty acids of 18C) [56]. Recently, Ruiz et al. [57] used a blend of bile salts as a dietary supplement to enhance the farming and production of Sparus aurata juveniles without compromising their intestinal health in terms of gut immune response and microbiota composition. Adding bile acid/salts in fish food can also mitigate the negative effect caused by partially or completely substituting fishmeal with alternative sources to increase protein and lipids (e.g., vegetables, rendering of marine and terrestrial animals). These dietary fishmeal alternatives often disturb the bile acid status in fish, resulting in either increased excretion/decreased intestinal reabsorption and/or decreased bile acid synthesis because of saponins, oligosaccharides, high-molecular-mass proteins, or other molecules that are believed to be involved in altering bile acid status [58].
The fact that four of the five fish extracts tested against p-NP substrates of different chain lengths showed high activities for p-NPC (8C; Figure 3), concerning p-NPM (14C) and p-NPP (16C), agrees with the result reported by Olsen et al. [59]. These authors observed that lipolytic activity in the pyloric ceca and intestinal tract of arctic charr hydrolyzes polyunsaturated and medium-chain saturated (like p-NPC) fatty acids better than longer-chain saturated and monounsaturated fatty acids.
The release of products via lipase hydrolysis increases with higher calcium levels, which means calcium controls the digestibility in the gastrointestinal tract. Hu et al. [60] found that the rate of fatty acid production increased with increasing calcium, e.g., the free fatty acids released after 20 min digestion were <12% for 0 mM of CaCl2 but >95% for 20 mM CaCl2. Contrary to this, lower activity can be observed if calcium-binding agents, such as EDTA, are present in the reaction system, because they are calcium-chelating agents.
The activation of p-NPM hydrolysis by Ca2+ (Figure 4) agrees with the lipase from rainbow trout, which requires mM concentrations of Ca2+ to hydrolyze triolein [56]. Calcium ions were necessary for the optimum activity of pyloric caeca lipases from Chinook salmon (Oncorhynchus tshawytscha) and hoki (Macruronus novaezelandiae) against caprate and palmitate esters of p-nitrophenol [61]. The activator effect of Ca2+ could be due to increased vmax and/or decreased KM [16]. Some ions (K+, Li+, Ni+, Co2+, Zn2+, Mg2+, Sn2+, Cu2+, Ba2+, Ca2+, and Fe2+) produce opposite results with different lipases [19,28]. In this sense, (phospho) lipases from the sea anemone Stichodactyla helianthus, selectively immobilized on octyl-Sepharose CL 4B support, showed an absolute requirement of Ca2+ to hydrolyze p-NP acetate [3]. However, a CEL from the hepatopancreas of red sea bream is Ca2+-independent [34].
Natural lipid fatty acids released by lipase activity are ionized in an aqueous solution by the loss of protons, and the reaction mixture tends to become acidic. The velocity of fatty acid release is proportional to the rate of proton release and, therefore, to the medium acidification speed. If NaOH is added at the same velocity when protons are released, the reaction mixture pH remains constant. The pH Stat assay consists of registering the velocity needed to add the titrant NaOH to the reaction mixture to maintain the pH at a fixed value. This velocity is proportional to the speed of enzymatic hydrolysis of substrate ester bonds, and therefore, its assessment allows for the determination of enzyme activity [23,51]. The pH Stat technique has been used to quantify lipase activity in plants, serum, plasma, and pancreas duodenal secretion, among other samples. The assay exhibits a detection limit of 0.1 μmol min−1. It can only be used in a restricted range of pH values, which should be equal to or higher than the apparent pKa of the released fatty acids, since these should be partially ionized [62].
Four different fish species were able to hydrolyze olive oil emulsions, as expected for the presence of true lipases in animal extracts (Figure 5). In addition, the in vitro oil digestibility of these species was positive for oils of olive, krill, and salmon. The digestibility of the fish oil was higher, as expected (Figure 6). However, conventional fish oil can be completely replaced in juvenile farmed salmon feeds by oil from the microorganism Schizochytrium sp. (T18), with no negative effects on the digestibility of dietary proximate nutrients, energy, or fatty acids [63].
Native protein electrophoresis and zymography are proposed to identify protein bands with lipase activity in fish samples. The quantities of lipase activity units in new studies of fish extracts could require adjustments to the amount of CE used in electrophoresis. The apparent sizes determined for the lipases from the five fish species (Figure 7) in this study are like those of the CELs from the hepatopancreas of red sea bream (64 kDa) [34], a tilapia lipase (46 kDa) [64], the pancreatic rainbow trout lipase (57 kDa) [56], and salmon (54.9 kDa) and hoki lipases (44.6 kDa) [13,61].
As mentioned above, fish lipase protocols vary in terms of substrates and physical conditions. Still, optimal pH must be measured in general terms, such as the pH from the intestine of the target fish, before the lipase assay. In marine fishes such as Thunnus orientalis, Totoaba macdonaldi, and Morone saxatils, the maximum lipolytic activity from pancreatic crude extracts was reported at pH 8.0, with temperatures ranging from 35 to 45 °C [65]. The highest lipolytic activity was observed at 35 °C and at a pH of 8–8.5 for Oncorhynchus tshawytscha [61]. However, the former authors did not measure the pH of the organs before the fish dissection.
Standard curves should be built at the same pH as activity assays because light absorption changes the function of pH due to variations in the ionization state. p-NP exhibits an absorption maximum at 400 nm at pH 8.0 (the pH value we recommend for activity assays). In a similar mode, a β-N derivative shows an absorption maximum at 540 nm at pH 9.0 [66]. The construction of standard curves in the pH range 6.0–10.0 (Supplementary Figure S5) agrees with the optimal pH for lipases (i.e., 7–9) in the neutral–basic zone, as has been observed in fish lumen where ranges oscillate at 6–9 [13,52,67,68,69]. Regarding pH extraction buffer and lipase assays, it is recommended that the pH be kept at 7.0–9.0, or, better, the assay could be undertaken in distilled water if the intestinal pH is unknown or if the optimum pH of fish lipases is expected to be determined [45]. On the other hand, we recommend activity assays at 25 °C. This is consistent with the work of Concha-Frías et al. [70], who reported an optimal temperature and pH of 35 °C and 9.0, respectively, for digestive lipase from the common snook (Centropomus undecimalis). Still, the assay is stable between 25 and 35 °C, and pH 5.0 and 8.0. Salmon and hoki lipases show an optimum pH of 8.0–8.5 [61]. A lipase from the alimentary canal and digestive gut of Catla catla exhibits an optimum pH and temperature of 7.8 and 20 °C, respectively [71].
5. Conclusions
In this work, we propose three methods to determine lipase activity. Two of them were based on spectrophotometric measurements following the hydrolysis of the chromogenic substrates p-NPM and β-NC. The other method, pH Stat, uses olive oil as a natural substrate of these enzymes to confirm lipase activity. It is important to analyze different sections of the intestine (e.g., anterior, medium, and posterior) considering the following recommendations for optimizing the experimental techniques used in this study: (a) The chain length of synthetic substrates (Figure 2 and Figure 3): 8C fatty acid (caprylic acid) chain length as a substrate (βNC) for non-bile salt-activated lipases and 14C fatty acid (stearic acid) chain length as a substrate (p-NPM) for bile salt-activated lipases. (b) The wavelength used in spectroscopic assessments (Supplementary Figures S1 and S3): 400 nm for p-NP and 540 nm for β-N. (c) Calcium concentration (Figure 4): 4 mM calcium chloride (Ca+2) final concentration in the reaction mixture. (d) The type (Figure 1) and concentration (Figure 2) of bile salt: 10 mg ml−1 NaT. (e) The experimental pH (according to fish intestinal pH): pH 8 using 20–30 mM Tris-HCl buffer. (f) The definition of enzymatic activity unit (one unit of lipase activity is defined as the enzyme amount that generates 1 μmol of β-N, p-NP, or fatty acid released per minute using the correct ε and the construction of standard curves) (Supplementary Figures S2 and S4): y = 16,876x for p-NP, and y = 24,286x for βN. This optimization could significantly improve comparisons among different studies. Finally, native protein electrophoresis and zymography are proposed to determine protein composition and lipase activity in fish samples (Figure 7).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes9070261/s1, Figure S1: Absorption spectra for p-NP and p-NPM. (a) p-NP (in DMSO) absorption spectra. (b) p-NP (E) and p-NPM (B) (in the final reaction mixture) absorption spectra; Figure S2: p-NP standard curve at pH 8.0 and 400 nm; Figure S3: Absorption spectra for β-N and β-NC. (a). β-N (in DMSO) absorption spectra. (b). β-N (E) and β-NC (B) (in the final reaction mixture) absorption spectra; Figure S4: β-N standard curve at pH 8.0 and 540 (a) and 550 (b) nm. U: universal buffer; Figure S6: Effect of pH on the slope of β-N standard curves.
Author Contributions
Conceptualization, H.N.-S.; methodology, H.N.-S. and C.A.A.-G.; formal analysis, H.N.-S. and D.T.-R.; investigation, H.N.-S. and C.A.A.-G.; resources, H.N.-S., D.T.-R. and C.A.A.-G.; writing—original draft preparation, H.N.-S., D.T.-R., C.A.A.-G., J.G.-B. and A.d.M.-M.; writing—review and editing, H.N.-S., F.V.-V., D.T.-R. and C.A.A.-G.; visualization, H.N.-S., F.V.-V. and C.A.A.-G.; supervision, H.N.-S., D.T.-R., C.A.A.-G., F.V.-V., J.G.-B. and A.d.M.-M.; project administration, H.N.-S. and D.T.-R.; funding acquisition, H.N.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financed by the CONAHCYT project 3266032 through the FORDECYT-PRONACES/2020 fund, which includes the design of enzymatic methods for the pygmy octopus. Thanks to CYTED Network LARVAplus (ref. 117RT0521).
Institutional Review Board Statement
The experiment on fishes did not need specific ethical approval in Mexico. The weakfish (Cynoscion parvipinnis), machete (Elops affinis), and rough triggerfish (Canthidermis maculata) were obtained from local fishermen, and the longfin yellowtail (Seriola rivoliana) and white snook (Centropomus viridis) were donated from one aquaculture company. Therefore, no national permit is required to harvest them; they are not endangered species.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available upon request from the authors.
Acknowledgments
The authors thank (in memorial) Patricia Hinojosa BaltazarϮ (lab work) from CIBNOR for her technical support to Luis Javier Nolasco-Alzaga in statistical analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- De Caro, J.D.; Rouimi, P.; Rovery, M. Hydrolysis of p-nitrophenyl acetate by the peptide chain fragment (336–449) of porcine pancreatic lipase. Eur. J. Biochem. 1986, 158, 601–607. [Google Scholar] [CrossRef] [PubMed]
- del Monte, A.; González-Bacerio, J.; Romero, L.; Aragón, C.; Vega, F.; Nolasco, H.; Díaz, J. Extraction systems for isolating esterases having interfacial adsorption. Rev. Colomb. Biotecnol. 2009, 11, 7–16. Available online: http://scielo.org.co/pdf/biote/v11n1/v11n1a02.pdf (accessed on 18 January 2024).
- del Monte-Martínez, A.; González-Bacerio, J.; Aragón-Abreu, C.; Palomo-Carmona, J.M.; Guisán-Seijas, J.M.; Díaz-Brito, J. Selective and oriented immobilization of (phospho)lipases from the Caribbean Sea anemone Stichodactyla helianthus (Ellis, 1768) by interfacial adsorption. Rev. CENIC Cienc. Biológicas 2012, 43, 3–8. Available online: https://doaj.org/article/5b82bf6757e54104bb9125eb1e507aae (accessed on 15 March 2024).
- del Monte-Martínez, A.; González-Bacerio, J.; Cutiño-Avila, B.; Ruiz, R.; Avila, R.; Ramos-Leal, M.; Nolasco, H.; Díaz, J.; Guisán, J.M. Esters biotransformation by immobilized interfacial esterases from the Caribbean Sea anemone Stichodactyla helianthus. Biotecnol. Apl. 2015, 32, 3201–3210. Available online: https://doaj.org/article/848e3ddee2bb40d58171eba59562930a (accessed on 15 March 2024).
- del Monte-Martínez, A.; González-Bacerio, J.; Varela, C.M.; Vega-Villasante, F.; Lalana-Rueda, R.; Nolasco, H.; Díaz, J.; Guisán, J.M. Screening and immobilization of interfacial esterases from marine invertebrates as promising biocatalyst derivatives. Appl. Biochem. Biotechnol. 2019, 189, 903–918. [Google Scholar] [CrossRef]
- Rúa, M.L.; Diaz-Maurino, T.; Fernadez, V.M.; Otero, C.; Ballesteros, A. Purification and characterization of two distinct lipases from Candida cylindracea. Biochim. Biophys. Acta 1993, 1156, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Dosanjh, N.S.; Kaur, J. Biochemical analysis of a native and proteolytic fragment of a high-molecular-weight thermostable lipase from a mesophilic Bacillus sp. Prot. Express Purif. 2002, 24, 71–75. [Google Scholar] [CrossRef]
- Yúfera, M.; Moyano, F.J.; Martínez-Rodríguez, G. The digestive function in developing fish larvae and fry. From molecular gene expression to enzymatic activity. In Emerging Issues in Fish Larvae Research; Yúfera, M., Ed.; Springer International Publishing AG: Cham, Switzerland, 2018; p. 296. [Google Scholar] [CrossRef]
- Dandavate, V.; Jinjala, J.; Keharia, H.; Madamwar, D. Production, partial purification and characterization of organic solvent tolerant lipase from Burkholderia multivorans V2 and its application for ester synthesis. Bioresour. Technol. 2009, 100, 3374–3381. [Google Scholar] [CrossRef]
- De Caro, J.D.; Chautan, M.P.; Rouimi, P.; Rovery, M. Acetylation of Lys-373 in porcine pancreatic lipase after reaction of the enzyme or its C-terminal with p-nitrophenyl acetate. Biochimie 1988, 70, 1785–1790. [Google Scholar] [CrossRef]
- Gupta, R.; Rath, P.; Gupta, N.; Bradoo, S. Lipase assays for conventional and molecular screening: An overview. Biotech. Appl. Biochem. 2003, 37, 63–71. [Google Scholar] [CrossRef]
- Lopez-Amaya, C.; Marangoni, A.G. Lipases. In Seafood Enzymes. Utilization and Influence on Postharvest Seafood Quality; Haard, N.F., Simpson, B.K., Eds.; Marcel Dekker: New York, NY, USA, 2000; pp. 121–146. [Google Scholar] [CrossRef]
- Kurtovic, I.; Marshall, S.N.; Zhao, X.; Simpson, B.K. Lipases from mammals and fishes. Rev. Fish. Sci. 2009, 17, 18–40. [Google Scholar] [CrossRef]
- Aloulou, A.; Puccinelli, D.; De Caro, A.M.; Leblond, Y.; Carriere, F. A comparative study on two fungal lipases from Thermomyces lanuginosus and Yarrowia lipolytica shows the combined effects of detergentsand pH on lipase adsorption and activity. Biochim. Biophys. Acta 2007, 1771, 1446–1456. [Google Scholar] [CrossRef] [PubMed]
- Sonesson, A.W.; Elofsson, U.M.; Brismar, H.; Callisen, T.H. Adsorption and mobility of a lipase at a hydrophobic surface in the presence of surfactants. Langmuir 2006, 22, 5810–5817. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Futami, Y.; Tarui, S.; Shinomiya, T. Activation of human pancreatic lipase activity by calcium and bile salts. J. Biochem. 1982, 92, 243–251. [Google Scholar] [CrossRef]
- Larsson, A.; Erlanson-Albertsson, C. The importance of bile salt for the reactivation of pancreatic lipase by colipase. Biochim. Biophys. Acta 1983, 750, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Barnescu, R.; Serban, M.; Rugina, A.; Crisan, I.; Cepisca, C.; Caloianu, M. Biochemical characteristics in heterogeneous catalysis correlated with bioactive effect of the extracted components. Rom. J. Biol. Sci. 1997, 1–2, 84–89. [Google Scholar]
- Wu, X.Y.; Jaaskelainen, S.; Linko, Y.Y. Purification and partial characterization of Rhizomucor miehei lipase for ester synthesis. Appl. Biochem. Biotech. 1996, 59, 145–158. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fuentes, M.; Fernandez-Lorente, G.; Mateo, C.; Guisan, J.M.; Fernandez-Lafuente, R. General trend of lipase to self-assemble giving bimolecular aggregates greatly modifies the enzyme functionality. Biomacromolecules 2003, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Guisan, J.M.; Fernandez-Lafuente, R.; Bastida, A.; Blanco, R.M.; Soler, G.; Garcia-Junceda, E. Modulation of activity/stability properties of lipase from Pseudomonas flourescens by multipoint covalent immobilization on glyoxyl-supports. In Engineering of/with Lipases; Malcata, F.X., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1996; pp. 243–256. Available online: http://tdl.libra.titech.ac.jp/journaldocs/recordID/article.bib-01/ZR000000015830 (accessed on 3 February 2024).
- Sharma, A.K.; Tiwari, R.P.; Hoondal, G.S. Properties of a thermostable and solvent stable extracellular lipase from a Pseudomonas sp. AG-8. Biotechnol. Bioeng. 2002, 77, 693–703. [Google Scholar] [CrossRef]
- Nolasco-Soria, H.; Moyano-López, F.; Vega-Villasante, F.; Del Monte, A.; Espinoza-Chaurand, D.; Gisbert, E. Lipase and Phospholipase Activity Methods for Marine Organisms. In Lipases and Phospholipases: Methods and Protocols; Methods in Molecular Biology; Sandoval, G., Ed.; Humana Press: New York, NY, USA, 2018; Chapter 7; Volume 1835. [Google Scholar] [CrossRef]
- Nolasco-Soria, H. Fish digestive lipase quantification methods used in aquaculture studies. Front. Aquac. 2023, 2, 21225216. [Google Scholar] [CrossRef]
- Bornscheuer, U.; Reif, O.W.; Lausch, R.; Freitag, R.; Scheper, T.; Kolisis, F.N.; Menge, U. Lipase of Pseudomonas cepacia for biotechnological purposes: Purification, crystallization and characterization. Biochim. Biophys. Acta 1994, 1201, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zeng, Q.M.; Zong, M.H. Substrate specificity of lipase from Burkholderia cepacia in the synthesis of 3-arylaliphatic acid esters of floxuridine. J. Biotechnol. 2009, 142, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Berner, D.L.; Hammond, E.G. Phylogeny of lipase specificity. Lipids 1970, 5, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Snellman, E.A.; Sullivan, E.R.; Colwell, R.R. Purification and properties of the extracellular lipase, LipA, of Acinetobacter sp. RAG-1. Eur. J. Biochem. 2002, 269, 5771–5779. [Google Scholar] [CrossRef] [PubMed]
- Zouari, N.; Miled, N.; Cherif, S.; Mejdoub, H.; Gargouri, Y. Purification and characterization of a novel lipase from the digestive glands of a primitive animal: The scorpion. Biochim. Biophys. Acta 2005, 1726, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Cygler, M.; Grochulski, P.; Schrag, J.D. Structural determinants defining common stereoselectivity of lipases toward secondary alcohols. Can. J. Microb. 1995, 41, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T. Methods to increase enantioselectivity of lipases and esterases. Curr. Opin. Biotech. 2002, 13, 543–547. [Google Scholar] [CrossRef]
- Jensen, R.G.; De Jong, F.A.; Lambert-Davis, L.G.; Hamosh, M. Fatty acid and positional selectivities of gastric lipase from premature human infants, in vitro studies. Lipids 1994, 29, 433–435. [Google Scholar] [CrossRef] [PubMed]
- González-Bacerio, J.; Rodríguez, J.; del Monte, A. Lipases: Enzymes having the potential for developing immobilised biocatalysts by interfacial adsorption. Rev. Colomb. Biotecnol. 2010, 12, 124–140. Available online: http://www.scielo.org.co/scielo.php?pid=S0123-34752010000100013&script=sci_abstract#:~:text=Lipases%3A%20enzymes%20having%20the%20potential%20for%20developing%20immobilised,colomb.%20biotecnol%5Bonline%5D.%202010%2C%20vol.12%2C%20n.1%2C%20pp.113-140.%20ISSN%200123-3475 (accessed on 15 March 2024).
- Iijima, N.; Tanaka, S.; Ota, Y. Purification and characterization of bile salt-activated lipase from the hepatopancreas of red sea bream, Pagrus major. Fish. Physiol. Biochem. 1998, 18, 59–69. [Google Scholar] [CrossRef]
- Versaw, W.K.; Cuppett, S.L.; Winters, D.D.; Williams, L.E. An improved colorimetric assay for bacterial lipase in nonfat dry milk. J. Food Sci. 1989, 54, 1557–1558. [Google Scholar] [CrossRef]
- Bier, M. Lipases. In Methods in Enzymology; Colowick, S.P., Kaplan, N.O., Eds.; Academic Press: New York, NY, USA, 1955; Volume 1, pp. 627–642. [Google Scholar]
- Bertolini, M.C.; Schrag, J.D.; Cygler, M.; Ziomek, E.; Thomas, D.Y.; Vernet, T. Expression and Characterization of Geotrichum candidum Lipase I Gene. FEBS J. 1995, 228, 863–869. [Google Scholar] [CrossRef]
- Sastry, K.V.; Gupta, P.K. In vitro inhibition of digestive enzymes by heavy metals and their reversal by chelating agent: Part I. Mercuric chloride intoxication. Bull. Environ. Contam. Toxicol. 1978, 20, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Tyndall, J.D.; Sinchaikul, S.; Fothergill-Gilmore, L.A.; Taylor, P.; Walkinshaw, M.D. Crystal structure of a thermostable lipase from Bacillus stearothermophilus P1. J. Mol. Biol. 2002, 323, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Amada, K.; Kwon, H.J.; Haruki, M.; Morikawa, M.; Kanaya, S. Ca2+-induced folding of a family I.3 lipase with repetitive Ca2+ binding motifs at the C-terminus. FEBS Lett. 2001, 509, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Egmond, M.R. Action of lipases. In Engineering of/with Lipases; Malcata, F.X., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1996; pp. 183–191. Available online: https://api.semanticscholar.org/CorpusID:99992337 (accessed on 20 February 2024).
- Tsai, S.W.; Lu, C.C.; Chang, C.S. Surfactant enhancement of (S)-naproxen ester productivity from racemic naproxen by lipase in isooctane. Biotech. Bioeng. 1996, 51, 148–156. [Google Scholar] [CrossRef]
- Solovyev, M.M.; Izvekova, G.I. Seasonal changes in pH values in the intestine of fish from Lake Chany (West Siberia). Inland Water Biol. 2016, 9, 400–404. [Google Scholar] [CrossRef]
- Gisbert, E.; Nolasco, H.; Solovyev, M. Towards the standardization of brush border purification and intestinal alkaline phosphatase quantification in fish with notes on other digestive enzymes. Aquaculture 2018, 487, 102–108. [Google Scholar] [CrossRef]
- Nolasco-Soria, H. Improving and standardizing protocols for alkaline protease quantification in fish. Rev. Aquac. 2020, 13, 43–65. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Nolasco-Soria, H. Amylase quantification in aquaculture fish studies: A revision of most used procedures and presentation of a new practical protocol for its assessment. Aquaculture 2021, 538, 736536. [Google Scholar] [CrossRef]
- Meier, A.R.; Yehl, J.B.; Eckenroad, K.W.; Manley, G.A.; Strein, T.G.; Rovnyak, D. Stepwise aggregation of cholate and deoxycholate dictates the formation and loss of surface-available chirally selective binding sites. Langmuir 2018, 34, 6489–6501. [Google Scholar] [CrossRef] [PubMed]
- Campanelli, A.R.; Candeloro De Sanctis, F.; D’Archivio, A.A.; Giglio, E.; Scarammuzza, L. Crystal structures of bile salts: Sodium taurocholate. J. Incl. Phenom. Macrocycl. Chem. 1991, 11, 247–256. [Google Scholar] [CrossRef]
- Cherry, I.S.; Crandall, L.A. The specificity of pancreatic lipase: Its appearance in the blood after pancreatic injury. Am. J. Physiol. 1932, 100, 266–273. [Google Scholar] [CrossRef]
- Nolasco, H. Métodos Utilizados por el Centro de Investigaciones Biológicas del Noroeste (CIBNOR) para la Medición de Digestibilidad in vitro para Camarón. In Manual de Metodologías de Digestibilidad In Vivo e In Vitro Para Ingredientes y Dietas Para Camarón; Cruz Suárez, L.E., Villarreal Colmenares, H., Salazar, M.T., Nieto López, M.G., Villarreal Cavazos, D.A., Ricque Marie, D., Eds.; Universidad Autónoma de Nuevo León: Monterrey, Mexico, 2008; pp. 215–225. Available online: https://nutricionacuicola.uanl.mx/public/site/images/admin/manual_metodologias.pdf (accessed on 9 January 2024).
- Rasco, B.A.; Hultin, H.O. A comparison of dogfish and porcine pancreatic lipases. Comp. Biochem. Physiol. B 1988, 89, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Løkka, G.; Dhanasiri, A.K.S.; Krogdahl, A.; Kortner, T.M. Bile components affect the functions and transcriptome of the rainbow trout intestinal epithelial cell line RTgutGC. Fish. Shellfish. Immun. 2022, 131, 1144–1156. [Google Scholar] [CrossRef]
- Gao, Y.; Yao, Y.; Huang, J.; Sun, Y.; Wu, Q.; Guo, D.; Wang, S. Effect of dietary bile acids supplementation on growth performance, feed utilization, intestinal digestive enzyme activity and fatty acid transporters gene expression in juvenile leopard coral grouper (Plectropomus leopardus). Front. Mar. Sci. 2023, 10, 1171344. [Google Scholar] [CrossRef]
- Hui, D.Y.; Howles, P.N. Carboxyl ester lipase: Structure-function relationship and physiological role in lipoprotein metabolism and atherosclerosis. J. Lipid Res. 2002, 43, 2017–2030. [Google Scholar] [CrossRef]
- Leger, C.; Bauchart, D.; Flanzy, J. Some properties of pancreatic lipase in Salmo gairdnerii Rich.: Km, effects of bile salts and Ca2+, gel filtrations. Comp. Biochem. Physiol. B 1977, 57, 359–363. [Google Scholar] [CrossRef]
- Ruiz, A.; Andree, K.B.; Furones, D.; Holhorea, P.G.; Calduch-Giner, J.À.; Viñas, M.; Pérez-Sánchez, J.; Gisbert, E. Modulation of gut microbiota and intestinal immunenresponse in gilthead seabream (Sparus aurata) by dietary bile salt supplementation. Front. Microbiol. 2023, 14, 1123716. [Google Scholar] [CrossRef]
- Romano, N.; Kumar, V.; Yang, G.; Kajbaf, K.; Rubio, M.B.; Overturf, K.; Brezas, A.; Hardy, R. Bile acid metabolism in fish: Disturbances caused by fishmeal alternatives and some mitigating effects from dietary bile inclusions. Rev. Aquac. 2020, 12, 1792–1817. [Google Scholar] [CrossRef]
- Olsen, R.E.; Henderson, R.J.; Ringo, E. The digestion and selective absorption of dietary fatty acids in Arctic charr, Salvelinus alpinus. Aquac. Nutr. 1998, 4, 13–21. [Google Scholar] [CrossRef]
- Hu, M.; Li, Y.; Decker, E.A.; McClements, D.J. Role of calcium and calcium-binding agents on the lipase digestibility of emulsified lipids using an in vitro digestion model. Food Hydrocoll. 2010, 24, 719–725. [Google Scholar] [CrossRef]
- Kurtovic, I.; Marshall, S.N.; Zhao, X.; Simpson, B.K. Purification and properties of digestive lipases from Chinook salmon (Oncorhynchus tshawytscha) and New Zealand hoki (Macruronus novaezelandiae). Fish. Physiol. Biochem. 2010, 36, 1041–1060. [Google Scholar] [CrossRef] [PubMed]
- Beisson, F.; Tiss, A.; Riviere, C.; Verger, R. Methods for lipase detection and assay: A critical review. Eur. J. Lipid Sci. Technol. 2000, 102, 133–153. [Google Scholar] [CrossRef]
- Tibbetts, S.M.; Scaife, M.A.; Armenta, R.E. Apparent digestibility of proximate nutrients, energy and fatty acids in nutritionally-balanced diets with partial or complete replacement of dietary fish oil with microbial oil from a novel Schizochytrium sp. (T18) by juvenile Atlantic salmon (Salmo salar L.). Aquaculture 2020, 520, 735003. [Google Scholar] [CrossRef]
- Taniguchi, A.; Takano, K.; Kamoi, I. Purification and properties of lipase from Tilapia intestine-digestive enzyme of Tilapia-VI. Nippon Suisan Gakkaishi 2001, 67, 78–84. [Google Scholar] [CrossRef]
- Rueda-Lopez, S.; Martinez-Montano, E.; Viana, M.T. Biochemical characterization and comparison of pancreatic lipases from the Pacific bluefin tuna, Thunnus orientalis; totoaba, Totoaba macdonaldi; and striped bass, Morone saxatilis. J. World Aquac. Soc. 2017, 48, 156–165. [Google Scholar] [CrossRef]
- Espada, J.; Horobin, R.W.; Stockert, J.C. Fluorescent cytochemistry of acid phosphatase and demonstration of fluid-phase endocytosis using an azo dye method. Histochem. Cell Biol. 1997, 108, 481–487. [Google Scholar] [CrossRef]
- Mukundan, M.K.; Gopakumar, K.; Nair, M.R. Purification of a lipase from the hepatopancreas of oil sardine (Sardinella longiceps Linnaeus) and its characteristics and properties. J. Sci. Food Agric. 1985, 36, 191–203. [Google Scholar] [CrossRef]
- Borlongan, I.G. Studies on the digestive lipases of milkfish, Chanos chanos. Aquaculture 1990, 89, 315–325. [Google Scholar] [CrossRef]
- Gjellesvik, D.R.; Lombardo, D.; Walther, B.T. Pancreatic bile salt dependent lipase from cod (Gadus morhua): Purification and properties. Biochim. Biophys. Acta Lipids Lipid Metab. 1992, 1124, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Concha-Frías, B.; Gaxiola-Cortes, M.G.; De la Cruz-Alvarado, F.J.; Jimenez Martinez, L.D.; Peña-Marin, E.S.; Oliva-Arriagada, M.A.; Arias-Moscoso, J.L.; Alvarez-González, C.A. Intestinal Lipase Characterization in Common Snook (Centropomus undecimalis) Juveniles. Fishes 2022, 7, 107. [Google Scholar] [CrossRef]
- Kameshwar Sharma, Y.V.R.; Neelima, B.; Prasidhi, T. Isolation, purification and characterization of secondary structure and kinetic study of lipase from Indian major carp, Catla catla (Catla). Enzym. Eng. 2014, 3, 121. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).