Abstract
Metschnikowia bicuspidata, a globally distributed opportunistic pathogenic fungus, poses a significant threat to crustaceans in diverse aquatic ecosystems, causing severe diseases. Iron, recognized as a virulence factor, plays a crucial role in successful infection with M. bicuspidata. Therefore, this study aims to investigate the transcriptome response of M. bicuspidata to low- and high-iron conditions. Overall, 1082 differentially expressed genes (DEGs) (FDR < 0.05, |log2FC ≥ 1.5|) were identified, comprising 977 and 105 DEGs, in response to low- and high-iron conditions, respectively. These genes predominantly participate in altering metabolism, cell membranes, or cellular structure, allowing the organism to adapt to varying iron levels. Iron limitation-induced genes play crucial roles in energy metabolism, transport, and catabolism pathways. Moreover, 27 ortholog genes were associated with iron transport and homeostasis, with 7 of them participating in iron uptake and regulation under low-iron conditions. This study contributes to the comprehension of iron homeostasis in aquatic fungi. It may offer potential therapeutic strategies for managing M. bicuspidata diseases.
1. Introduction
Metschnikowia bicuspidata, a yeast with opportunistic pathogenic characteristics, is globally distributed in marine and freshwater environments. Within the context of Daphnia–M. bicuspidata interactions, it serves as a well-studied organism for exploring host parasite theory []. Moreover, M. bicuspidata is characterized as an obligate killer, implying that once it infects the host, recovery is not possible []. M. bicuspidata spores are either directly consumed by aquatic hosts or indirectly ingested by diseased animals. It rapidly proliferates within the bloodstream of the host. Upon death, ascospores are released into the aquatic environment [,]. Therefore, many economically important aquatic species, such as the freshwater prawn Macrobrachium rosenbergii [], Chinese swimming crab Portunus Trituberculatus [], Chinese mitten crab Eriocheir sinensis [], Chinese grass shrimp Palaemonetes sinensis [], and chinook salmon [], have experienced high mortality rates due to M. bicuspidata infections, leading to a substantial decrease in production and considerable economic losses [].
Iron functions as a redox cofactor essential for various fundamental biological processes, such as cellular respiration, lipid biosynthesis, DNA replication and repair, ribosome biosynthesis, and circulation. Positioned as a crucial trace element at the interface between hosts and pathogens, iron plays a vital role in the survival, persistence, and virulence of microbial infections. During in vivo infection, fungi encounter significantly reduced levels of free iron in the bloodstream. To effectively proliferate and establish an infection within the host, extracellular fungi employ various strategies to break the host iron restriction mechanism. Consequently, the competition for iron between the pathogen and host emerges as a crucial determinant of the outcome of an infection. However, as commensals within the animal/human gastrointestinal tract, fungi encounter elevated levels of free iron. Iron overload leads to oxidative damage to cells through the Fenton reaction []. Therefore, fungal pathogens have evolved intricate iron homeostasis regulatory mechanisms. This includes a diverse array of iron acquisition systems aimed at efficiently extracting iron from the host, regulatory factors that modulate iron transport and storage, and associated metabolic processes to attain sufficient but non-toxic levels of this essential micronutrient []. Comprehensive insights into the regulation mechanisms of iron homeostasis in fungi have been most extensively studied in two model yeasts, Saccharomyces cerevisiae and Schizosaccharomyces pombe, as well as in the human pathogen Candida albicans [,,,,]. While detailed descriptions of iron regulation networks in the saprophytic filamentous ascomycete Aspergillus fumigatus [], basidiomycete Cryptococcus neoformans [], and Candida glabrata also exist [], these studies reveal that distinct molecular mechanisms regulate iron metabolism in each species, highlighting both common and species-specific mechanisms employed by fungi to acquire iron and regulate their response to iron conditions [,,]. Therefore, this study aims to explore how the transcriptome of aquatic pathogenic fungi, M. bicuspidata, responds to both low- and high-iron conditions, and we also obtained several key genes involved in iron transport and regulation. These findings contribute to advancing our understanding of iron homeostasis in aquatic fungi and offer potential therapeutic approaches for managing fungal diseases caused by M. bicuspidata.
2. Materials and Methods
2.1. Yeast Strains
The purified yeast M. bicuspidata strain LNES0119 isolated from Chinese mitten crabs in Panjin city (Liaoning Province, China) was cultured in solid YPD medium for 48 h at 28 °C. Subsequently, it was transferred to a fresh medium and cultured for an additional 48 h before yeast collection. To characterize the growth traits of the M. bicuspidata strains LNES0119 under different iron concentrations, the strains were aerobically cultured in 50 mL of liquid synthetic defined (SD) medium (SD: 0.67% yeast nitrogen base [YNB, free of iron, Coolaibo Technology Co., Ltd.-Beijing, China], 2% glucose, and 0.079% complete supplemental mixture [CSM; Formedium]). Various concentrations of ferrous ammonium sulfate were added. All cells were incubated at 28 °C, and the OD values at 600 nm were measured using a spectrophotometer. These data were used for further RNA sequencing sample collection.
2.2. Library Preparation and RNA Sequencing
Yeast strains were cultured overnight under three iron concentrations, 0 µM (iron-limited, L-Fe), 100 µM (iron-replete, N-Fe), or 500 µM (high-iron, H-Fe), reaching the mid-log phase (A600 of 0.4–0.6) in L-Fe, N-Fe, or H-Fe medium (three replicates for each sample). Under iron-deficient conditions (−Fe), the yeast was cultured in liquid SD medium with iron-free YNB. Iron-enriched conditions involved SD medium supplemented with ferrous ammonium sulfate. Subsequently, yeast cells were collected for RNA extraction and transcriptome sequencing. Overall, total RNA was extracted with a Takara RNAiso Plus kit (Takara, Shiga, Japan) following the manufacturer’s instructions. Then, 1 μg RNA per sample served as input material for the RNA sample preparations. RNA concentration, quality, and integrity were assessed with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing libraries were generated using the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, Waltham, MA, USA), following the instructions of the manufacturer. Index codes were incorporated to attribute sequences to each sample. Subsequently, PCR products were purified (using the AMPure XP system), and library quality was evaluated using the Agilent Bioanalyzer 2100 system. Index-coded samples were clustered using a cBot Cluster Generation System with the TruSeq PE Cluster Kit v4-cBot-HS (Illumina) according to the instructions of the manufacturer. Library preparations were conducted on an Illumina HiSeq platform, resulting in the generation of paired-end 150 bp reads.
2.3. RNA-Seq Data Analysis
Adapters of raw sequencing reads were eliminated using cutadapt v. 2.20 [], and fastp v0.20.1 [] with default parameters was employed to filter low-quality reads (average base mass value less than 20). The resulting clean reads were then mapped to the reference genome of Metschnikowia bicuspidata NRRL YB-4993 (PRJNA207846) (accession no. GCA_001664035.1) using HISAT2 v2.1.0 []. Clean high-quality reads from all samples were assembled using Trinity v. 2.10.0. [] to create a unified set of sequences for subsequent analysis. The assembled unigenes were aligned against NCBI non-redundant protein sequences (Nr) to identify the optimal transcript as a representative gene. Annotation of the possible pathways of the unigenes was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/, accessed on 4 December 2023) through BLASTP, with an e-value cutoff of 1e–5. Additionally, gene annotation based on gene ontology (GO) was conducted using BLAST 2GO [] (http://www.geneontology.org, accessed on 4 December 2023).
The gene expression level of each transcript was calculated using fragments per kilobase of transcript per million fragments mapped (FRKM). False discovery rate (FDR) correction was applied to the p values. Differentially expressed genes (DEGs) were identified to assess transcriptional changes in the yeast strain under different iron stresses by comparing the expression levels between the N-Fe vs. L-Fe and N-Fe vs. H-Fe groups. Genes exhibiting |log2FoldChange|≥1.5 and p < 0.05, identified by DEGs, were recognized as differentially expressed between the two treatments using DEGseq v. 1.42.0. Subsequently, GO and KEGG analyses were conducted on the DEGs.
2.4. Real-Time qRT-PCR Validation
To verify the RNA transcript data, 11 DEGs were examined by relative quantitative real-time PCR (RT-qPCR). Specific primers for qRT-PCR were designed with Premier Primer 5. Table S1 shows the sequences. RT-PCR was conducted as previously described [], with Actin serving as the reference gene for normalization. The gene expression levels from three biological replicates were analyzed using the 2−ΔΔCT method []. Data were reported as means ± standard errors (standard error of the mean, SEM).
2.5. Statistical Analyses
Raw experimental data were analyzed with Excel 2013 (Microsoft, Redmond, WA, USA). Significant differences were assessed using one-way ANOVA in SPSS software (version 17.0; SPSS Inc., Chicago, IL, USA) with Tukey’s multiple range test. The results are expressed as means ± standard deviations (SDs) and are visualized using GraphPad Prism 6.0 (GraphPad Software, La Jolla, LA, USA).
3. Results
3.1. Overviews of Yeast Cultivation and RNA-Seq Data Analysis
The M. bicuspidata strains LNES0119 were cultured under iron-deficient (0 µm, L-Fe group), iron-replete (100 µM, N-Fe), and iron-rich conditions (500 µm, H-Fe group), then the yeasts were sampled for RNA sequencing to evaluate the transcriptional response under various iron stresses. Nine cDNA libraries were employed for the transcriptome sequencing, yielding over 6.1 billion clean bases and 20.5 million clean reads from each library. The percentages of Q20 and Q30 were consistently higher than 98.1 and 93.17–94.68%, respectively (Table S2). Additionally, 86.55% to 87.55% of clean reads from each library were uniquely mapped to the reference genome.
3.2. Identification of Differentially Expressed Genes (DEGs) Related to Iron Stress
Differential expression analysis between the sample groups was conducted using DESeq2 to identify a set of DEGs. Overall, 977 DEGs were observed under low-iron conditions, with 605 upregulated and 372 downregulated. In contrast, only 105 DEGs were identified under high-iron conditions, with 72 upregulated and 33 downregulated. Additionally, 62 DEGs responded to low- and high-iron stress (Figure 1). Eleven genes related to iron homeostasis and iron metabolism, including Fet3, Sit1, Ccc1, Ftr1, Fre1, Sfu1, Cfl1, Grx5, Sdh2, Aco1, and Snf1, were selected in N-Fe versus L-Fe for qRT-PCR verification, and the expression data of these genes exhibited high coordination with the transcriptome data (Figure 2).
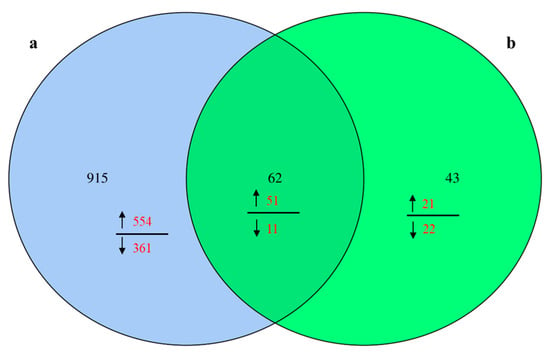
Figure 1.
Venn diagram illustrating the DEGs in iron-deficient (a, L-Fe vs. N-Fe) and -rich conditions (b, H-Fe vs. N-Fe). The upward arrow denotes upregulated gene expression, while the downward arrow indicates downregulated gene expression.
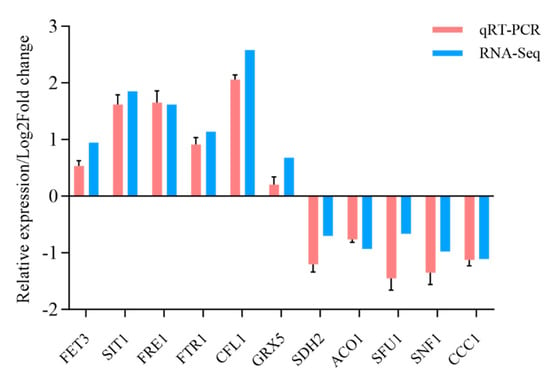
Figure 2.
Validation of differential gene expression profile through qRT-PCR. A total of 11 genes were validated using qRT-PCR against their expression profile from RNA-seq. The relative gene expression levels were quantitated using 2−ΔΔCt.
3.3. Gene Ontology (GO) Functional Annotation and Enrichment Analysis of the DEGs
GO enrichment analyses were conducted for the DEGs, revealing significant enrichment in biological processes (BPs), cellular components (CCs), and molecular functions (MFs) (Figure 3). Overall, the number of DEGs under low-iron stress surpassed those under high-iron stress in each classification, suggesting that the DEGs were predominantly associated with lower stress conditions. For molecular functions, the largest and second-largest groups were genes associated with catalytic activity (GO:0003824) and binding protein (GO:0005488) under low-iron stress. However, under high-iron stress, a reversal in protein function was observed. Analysis of the cellular components of the proteins encoded by the DEGs revealed that most proteins comprised integral/intrinsic membrane parts, including cells (GO:0005623), cell parts (GO:0044464), and organelles (GO:0043226), in response to iron stress. Regarding biological processes, the two largest groups were associated with metabolic processes (GO:0008152) and cellular processes (GO: GO:0009987) under both iron stress conditions. Overall, the results of the GO analysis suggest that M. bicuspidata might alter the metabolism and cell membrane or cellular structure in response to iron stress.
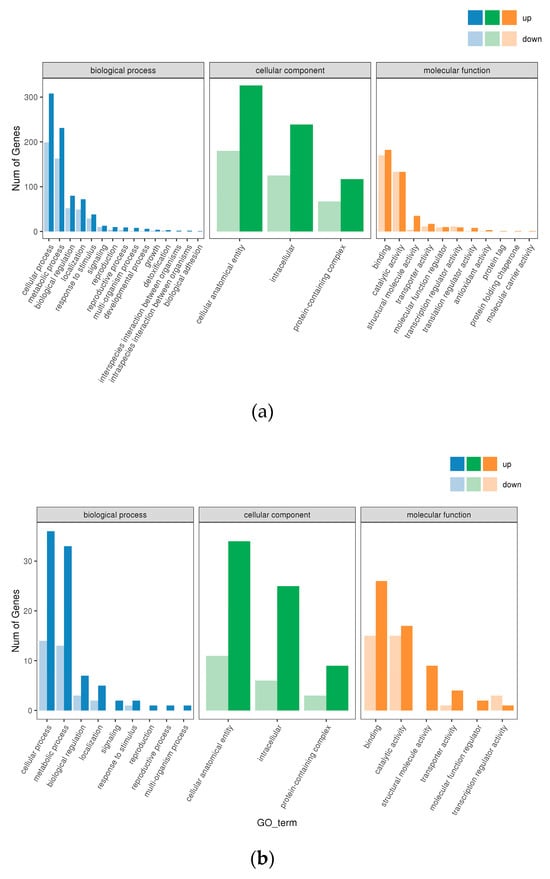
Figure 3.
Gene ontology (GO) annotation depicting DEGs in the (a) L-Fe vs. N-Fe and (b) H-Fe vs. N-Fe comparisons. Blue color represents the biological process (BP), green denotes the cellular component (CC), and orange represents the molecular function (MF).
DEGs under iron stress exhibited significant enrichment, categorized into 25 pathways, and belonging to four branches, namely cellular processes, environmental information processing, genetic information processing, and metabolism (Figure 4). Both low- and high-iron stress conditions showed enrichment in cell cycle–yeast (ko04111), meiosis–yeast (ko04113), MAPK signaling pathway–yeast (ko04011), ribosome (ko03010), RNA degradation (ko03018), ribosome biogenesis in eukaryotes (ko03008), RNA degradation (ko03018), and protein processing in the endoplasmic reticulum (ko04141) pathways. However, the number of DEGs under low-iron stress was considerably higher than that under high-iron stress. These pathways highlight the influence of iron on yeast cell growth, death, and genetic information processing. Moreover, under low-iron stress, the DEGs were primarily enriched in endocytosis (ko04144), autophagy-other (ko04136), oxidative phosphorylation (ko00190), as well as spliceosome (ko03040), ribosome biogenesis in eukaryotes (ko03008), mRNA surveillance (ko03015), RNA transport (ko03013), basal transcription factors (ko03022), biosynthesis of amino acids (ko01230), nucleotide excision repair (ko03420), purine metabolism (ko00230), carbon metabolism (ko01200), and citrate cycle (ko00020) pathways. These findings suggest that low iron levels may adversely affect energy metabolism, transport, and catabolism in yeast.
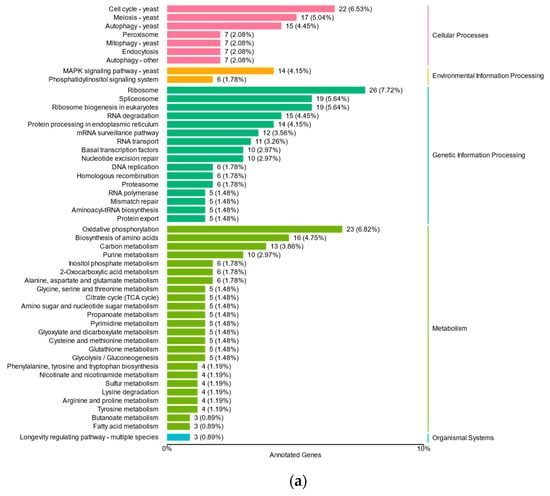
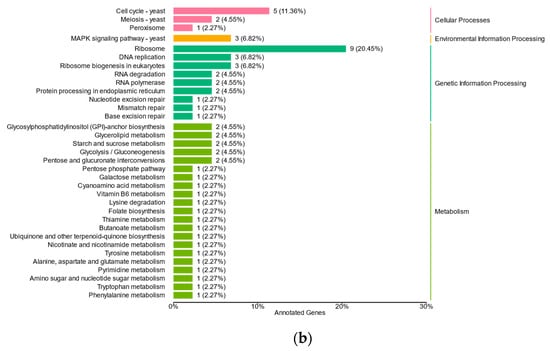
Figure 4.
Differentially expressed genes (DEGs) of M. bicuspidata under low- (a) and high-iron conditions (b) compared to those of normal iron fungus culturing. The distribution of these genes among the main functional categories is also illustrated.
3.4. DEGs Involved in MAPK Signaling Pathway in Iron Stress
According to KEGG enrichment analysis, the MAPK signaling pathway of M. bicuspidata exhibited enrichment with 14 DEGs under low-iron stress. Among these, Sst, Bem3, Cdc42, Ste18, Bnr1, Mss4, Pkc1, Hsl2, and Swe1 were upregulated, while Sac7, Paf1, Swi4, Ctt1, and Gre2 were downregulated. These genes predominantly participate in cell wall stress, high osmolarity, and pheromone processes. Conversely, only three upregulated DEGs, Swe1, Hsl1, and Sst2, were enriched in the MAPK signaling pathway under high-iron stress (Table 1) These signals were transmitted into the nucleus through the MAPK cascade, and under low-iron stress, Ctt1 and Gre2, which encode proteins involved in antioxidant stress, were found to be downregulated.

Table 1.
List of differentially expressed genes (DEGs) playing important roles in the MAPK signaling pathway. The log2FoldChange indicates the ratio of gene expression levels between the L-Fe vs. N-Fe comparisons (−Fe) and H-Fe vs. N-Fe comparisons (+Fe).
3.5. DEGs Involved in Oxidative Phosphorylation in Low-Iron Stress
The oxidative phosphorylation pathway was found to be enriched under low-iron stress. Within these pathways, 23 DEGs were identified, comprising 17 upregulated and 6 downregulated genes. Ten genes belonged to NADH reductase (Ndufs1, Ndufs4, Ndufs6, Ndufs7, Ndufv1, Ndufv2, Ndufa5, Ndufa12, Ndufb7, and Ndufb9) and four genes (QCR2, QCR7, QCR9, and QCR10) were associated with cytochrome reductase. Two genes (SDHD and SDHB) were related to fumarate reductase/succinate dehydrogenase. Three genes (COX4, COX6B, and COX7C) were part of cytochrome oxidase, while four genes were associated with ATP synthase (Table 2).

Table 2.
Differentially expressed genes (DEGs) associated with the oxidative phosphorylation pathway under low-iron stress. The log2FoldChange indicates the ratio of gene expression levels between the L-Fe vs. N-Fe comparisons.
3.6. DEGs Involved in Autophagy-Related Genes in Low-Iron Stress
Based on the KEGG enrichment analysis, 15 genes were enriched in the autophagy-yeast pathway, and six genes were enriched in the mitophagy pathway (Table 3). The autophagy-yeast genes included three (Sak1, ATG17, and Tap42) belonging to initiation, five (ATG6, ATG9, ATG11, and Ymr1) for nucleation, two (ATG18 and UME6) belonging to expansion, and three (VPS16, VPS33, and VPS41) belonging to the fusion process. In addition, the mitophagy pathway upregulated PKC1, UME6, MSS4, and MDM12 and downregulated CK2 and ATG11.

Table 3.
DEGs involved in autophagy-related genes under low-iron stress. The log2FoldChange indicates the ratio of gene expression levels between the between the L-Fe vs. N-Fe comparisons.
3.7. DEGs Involved in Iron Transport and Homeostasis
To enhance our understanding of iron transport and homeostasis in M. bicuspidata, BLAST analyses were conducted using the S. cerevisiae and C. albicans sequences based on the M. bicuspidata genome. The results showed a set of 27 genes encoding proteins involved in iron acquisition at the plasma membrane, iron import into the mitochondria, iron–sulfur cluster (ISC) biosynthesis, and vacuolar iron export (Table S3). Additionally, several key DEGs related to iron transport and homeostasis were identified as homologous genes in S. cerevisiae, C. albicans, A. fumigatus, and C. neoformans (Table 4). The results showed 10 putative genes characterized as ferric reductases (Table S3), possessing a ferric reductase domain and an FAD and/or NAD binding domain. Among these, three ferric reductase genes (Fre1, Cfl1) were upregulated under low stress. Multicopper ferroxidase (Fet3), iron permease (Ftr1), siderophore transport (Sit1), iron sensing, and transcription factors (Grx5) were upregulated under low stress. In contrast, the vacuolar Fe2+/Mn2+ transporter (Ccc1) gene, which exhibited downregulation under low-iron and upregulation under high-iron conditions, and the suppressor of ferric uptake 1 (Sfu1) were downregulated under low-iron conditions (Table 4).

Table 4.
Ortholog genes related to iron transport and homeostasis in M. bicuspidata and major human pathogenic fungi. The log2FoldChange indicates the ratio of gene expression levels between the L-Fe vs. N-Fe comparisons (-Fe) and H-Fe vs. N-Fe comparisons (+Fe). S. cerevisiae, C. albicans, A. fumigatus, and C. neoformans were human pathogenic fungi.
4. Discussion
Iron stress represents a prevalent form of nutrient stress faced by pathogens, playing a crucial role in infection and pathogenesis. The understanding of iron metabolism and homeostasis has been extensively established in model eukaryotes and other pathogenic fungi, each employing distinct strategies to adapt to varying intracellular iron levels. The yeast M. bicuspidata is a pathogen affecting aquatic animals of economic importance. Therefore, this study represents the first exploration of iron metabolism at the molecular level in aquatic pathogenic fungi, revealing numerous changes in low- and high-iron environments by transcript profile analysis.
Many research works have observed that the MAPK pathway plays crucial roles in cell cycle control, cell wall assembly and integrity, virulence, and responses to abiotic stress in fungi []. The majority of available information on fungal MAPK pathways is concentrated on S. cerevisiae and C. albicans. In this study, low iron and high iron both affected the MAPK pathway of M. bicuspidata. We observed the upregulation of genes involved in regulating the G2/M (Swe1 and Hsl1) and G1/S phase transitions (Swi4) [] under iron stress. Additionally, the expression of Cdc42, which regulates cell polarity, influencing morphology and growth in eukaryotes [], was demonstrated to be heightened in a low-iron environment in M. bicuspidata. These findings suggest that M. bicuspidata could be induced through the MAPK pathway, contributing to the regulation of bud growth and the cell cycle in response to iron stress. Additionally, the downregulation of Ctt1 and Gre2 in M., known to play roles in protecting cells against H2O2 stress in fungi [] under low iron levels, suggests a decrease in intracellular oxidative stress. Therefore, the MAPK pathway may be involved in iron homeostasis. In C. albicans, the MAPK pathway is known to contribute to iron homeostasis []. Furthermore, iron can induce the MAPK pathway, involving aspects such as cell wall architecture, cell surface flocculation, and adhesion in C. albicans [,,]. Therefore, the iron related to the MAPK pathway in M. bicuspidata should be well studied.
Oxidative phosphorylation (OXPHOS) is the predominant energy-producing pathway in eukaryotes, which involves coupling energy derived from the oxidation of specific metabolic substrates to the phosphorylation of adenosine bisphosphate (ADP), ultimately generating ATP. NADH reductase, the principal and first enzyme of the OXPHOS system, experiences evident inhibition of iron-dependent gene expression in M. bicuspidata under low-iron conditions. For example, Ndufs1, Ndufs7, Ndufv1, and Ndufv2, essential for electron transfer to ubiquinone [], are prominently affected. Conversely, an upregulation in other NADH reductase genes is observed, serving to protect the yeast and maintain subsequent metabolic processes. These findings are consistent with those observed in C. albicans cultivated under iron-deficient conditions [,]. Low iron levels can induce metabolic remodeling, including mitochondrial function and cellular energy metabolism in C. albicans, thereby sustaining normal intracellular ATP levels. Furthermore, S. cerevisiae cells under iron-deficient conditions demonstrate alterations in metabolic responses, including iron enzyme activities, amino acid homeostasis, and cellular membrane properties and functions []. These findings suggest that iron deficiency induces the oxidative phosphorylation pathway in M. bicuspidata through metabolic remodeling, ensuring an adequate energy supply.
Autophagy is a conserved intracellular degradation process designed to direct cytoplasmic material for lysosomal degradation and recycling, thereby maintaining cellular homeostasis. Extensive reviews on autophagy in filamentous fungi have highlighted its involvement in multiple physiological and pathological processes, including cell development, pathogenicity regulation, and programmed cell death [,,]. Fungal autophagy is commonly triggered by nutrient limitations (e.g., carbon, nitrogen, and metal ions). Autophagy functions as a recycling mechanism for nutrient or metal ion homeostasis, which is crucial for sustaining cell survival []. For example, the induction of general and mitochondrial autophagy in the human pathogenic yeast C. glabrata under iron-starved conditions contributes to longevity and pathogenicity []. Moreover, the iron regulon, CgAft1/2, has been identified as playing a role in activating autophagy genes []. Our study revealed 15 genes enriched in the autophagy-yeast pathway and six genes in the mitophagy pathway when M. bicuspidata was cultivated under iron-depleted conditions. These findings provide insight into the intimate connection between iron homeostasis and general and mitochondrial autophagy in M. bicuspidata, emphasizing the importance of further investigations in this domain.
Intracellular iron regulation in pathogenic fungi and model yeasts has been extensively investigated, leading to the identification of several iron homeostasis-related genes and transcription factors []. These control the uptake, utilization, and storage of iron in response to fluctuations in intracellular iron levels through distinct regulated network pathways. Fungi primarily employ either a high-affinity reductive iron acquisition (RIA) system or a siderophore uptake system to secure extracellular iron ions. The reductive iron uptake system is a highly conserved strategy among fungal species, such as S. cerevisiae, C. albicans, A. fumigatus, and C. neoformans. In this process, membrane ferric reductases (Fres) initially reduce Fe3+ to the more soluble Fe2+, followed by the re-oxidation of Fe2+ by multicopper ferroxidase (Fet3) to facilitate transportation through the iron permeability enzyme (Ftr1) [,]. However, ferric reductases exhibit variations among fungi, with a putative 8 in S. cerevisiae, 2 in S. prombe, 17 in C. albicans, 15 in A. fumigatus, 8 in C. neoformans, and 10 in M. bicuspidata in this study. Additionally, only 1–3 of the genes encoding ferric reductases exhibit cupric reductase activity and participate in iron acquisition []. In M. bicuspidata, three ferric reductase genes responded to low iron concentrations (Table 4; Figure 5). Another siderophore uptake system involves the absorption of exogenous iron-bound siderophores by other organisms through the siderophore transporter Sit/Arn in S. cerevisiae and Sit in C. albicans, A. fumigatus [], and M. bicuspidata. The imported Fe2+ is then transported intracellularly to the mitochondria or nucleus for energy metabolism, and excess iron ions are transported into the vacuoles by the Ccc1 transporter for storage, reuse, and detoxification (Figure 5). Iron homeostasis is regulated by iron-sensing transcription factors such as GATA- (SreA in A. fumigatus, Cir1 in C. neoformans, and Sfu1 in C. albicans) and CCAAT-type transcription factors (HapX in A. fumigatus and C. neoformans, and Hap43 in C. albicans) (Table 4). Iron-responsive GATA-type factors primarily control the expression of genes related to siderophore iron uptake, reductive iron uptake, and the mobilization of stored iron from vacuoles. Concurrently, CCAAT-type transcription factors play a role in repressing iron consumption while promoting the expression of iron uptake genes. These factors operate in a negative feedback loop and exhibit conservation across the fungal kingdom []. In C. albicans, the transcriptional factor CaSef1 interacts with the GATA-factor and CCAAT-factor core network, forming a regulatory circuit to adapt to diverse living niches []. Additionally, the model yeast S. cerevisiae features a distinctive system of Aft1/2 transcription factors that is not widely distributed []. Overall, fungi exhibit different and partially overlapping iron homeostasis regulation mechanisms, influenced by their diverse survival characteristics and the complexity of the ecological environment. In this study, we identified three homologous iron-sensing transcription factors (MbSfu1, MbHapX, and MbSef1) in M. bicuspidata (Table S3). Among them, only MbSfu1 was inhibited under low-iron stress (Table 4). Therefore, further investigation is needed to elucidate the regulatory mechanisms of these iron-sensing factors in M. bicuspidata.
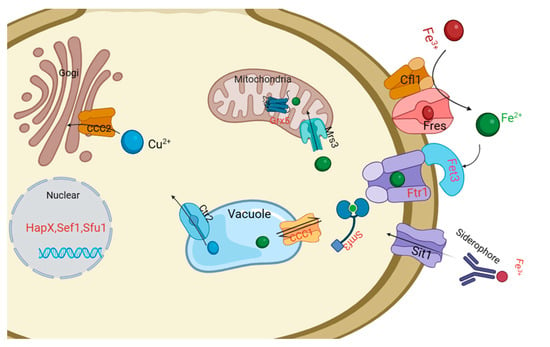
Figure 5.
Schematic diagram illustrating iron homeostasis in M. bicuspidata. Iron is taken up by a high-affinity reductive iron acquisition system consisting of ferric reductases (e.g., Fres and Clf1), the iron permease Ftr1, and multicopper ferroxidase Fet3. M. bicuspidata also transports iron through the siderophore transporter Sit1. The monothiol glutaredoxin Grx5 is presented as a putative sensor of iron–sulfur clusters and an interacting partner with transcription factors in the nucleus (HapX, Sef1, and Sfu1). The excess iron ions are transported into the vacuoles by the Ccc1 transporter for storage, reuse, and detoxification.
5. Conclusions
Our findings highlight the sensitivity of M. bicuspidata to low-iron conditions, showing enrichment in the MAPK, oxidative phosphorylation, and autophagy pathways. M. bicuspidata exhibits distinct iron-sensing and regulatory pathways in response to iron stress compared with other human pathogenic fungi. This prompts further investigation, suggesting a potential reference to the iron regulation modes of M. bicuspidata through human pathogenic fungi.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes9060236/s1, Table S1: Primers used for validation of differential gene expression. Table S2: Details of various samples taken for study and read sequence information. Table S3: Orthologs to genes related to iron transporter and homeostasis in M. bicuspidata and C. albicans.
Author Contributions
J.L., H.J. and J.B. conceived and designed the experiments as well as wrote the manuscript. Y.W., J.L., J.B. and S.Y. performed the experiments, data processing, analysis, and interpretation. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Liaoning Province Natural Science Foundation project (2022-MS-253); China Agriculture Research System of MOF and MARA (CARS-48); Liaoning Province “The Open Competition Mechanism to Select the Best Candidates” Project (2022JH1/10900005); Shenyang City “The Open Competition Mechanism to Select the Best Candidates” Project (22-316-2-01); Shenyang Science and Technology Mission Project (22-319-2-02); and Liaoning Province Department of Education fund (LJKQZ20222356).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data included in this study are available upon request by contacting the corresponding author.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Cáceres, C.E.; Tessier, A.J.; Duffy, M.A.; Hall, S.R. Disease in Freshwater Zooplankton: What Have We Learned and Where Are We Going? J. Plankton Res. 2014, 36, 326–333. [Google Scholar] [CrossRef]
- Ebert, D. Ecology, Epidemiology, and Evolution of Parasitism in Daphnia; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2005; ISBN 1-932811-06-0.
- Stewart Merrill, T.E.; Cáceres, C.E. Within-host Complexity of a Plankton-parasite Interaction. Ecology 2018, 99, 2864–2867. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Bao, J.; Cao, G.; Xing, Y.; Feng, C.; Hu, Q.; Li, X.; Chen, Q. Experimental Transmission of the Yeast, Metschnikowia bicuspidata, in the Chinese Mitten Crab, Eriocheir sinensis. J. Fungi 2022, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, T.; Wang, P.; Chen, Y.; Huang, J.; Lin, Y.; Chaung, H. Metschnikowia bicuspidata and Enterococcus faecium co-infection in the giant freshwater prawn Macrobrachium rosenbergii. Dis. Aquat. Org. 2003, 55, 161–167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, X.; Chi, Z.; Yue, L.; Li, J.; Li, M.; Wu, L. A Marine Killer Yeast against the Pathogenic Yeast Strain in Crab (Portunus trituberculatus) and an Optimization of the Toxin Production. Microbiol. Res. 2007, 162, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Jiang, H.; Shen, H.; Xing, Y.; Feng, C.; Li, X.; Chen, Q. First Description of Milky Disease in the Chinese Mitten Crab Eriocheir sinensis Caused by the Yeast Metschnikowia bicuspidata. Aquaculture 2021, 532, 735984. [Google Scholar] [CrossRef]
- Cao, G.; Bao, J.; Feng, C.; Li, X.; Lang, Y.; Xing, Y.; Jiang, H. First Report of Metschnikowia bicuspidata Infection in Chinese Grass Shrimp (Palaemonetes sinensis) in China. Transbounding Emerg. Dis 2022, 69, 3133–3141. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.M.; Strom, M.S. Infection and Mortality by the Yeast Metschnikowia bicuspidata Var. bicuspidata in Chinook Salmon Fed Live Adult Brine Shrimp (Artemia franciscana). Aquaculture 2003, 220, 43–57. [Google Scholar] [CrossRef]
- Chen, C.; Pande, K.; French, S.D.; Tuch, B.B.; Noble, S.M. An Iron Homeostasis Regulatory Circuit with Reciprocal Roles in Candida albicans Commensalism and Pathogenesis. Cell Host Microbe 2011, 10, 118–135. [Google Scholar] [CrossRef]
- Martínez-Pastor, M.T.; Puig, S. Adaptation to Iron Deficiency in Human Pathogenic Fungi. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2020, 1867, 118797. [Google Scholar] [CrossRef]
- Ramos-Alonso, L.; Romero, A.M.; Martínez-Pastor, M.T.; Puig, S. Iron Regulatory Mechanisms in Saccharomyces cerevisiae. Front. Microbiol. 2020, 11, 582830. [Google Scholar] [CrossRef] [PubMed]
- Labbé, S.; Pelletier, B.; Mercier, A. Iron Homeostasis in the Fission Yeast Schizosaccharomyces pombe. Biometals 2007, 20, 523. [Google Scholar] [CrossRef] [PubMed]
- Fourie, R.; Kuloyo, O.O.; Mochochoko, B.M.; Albertyn, J.; Pohl, C.H. Iron at the Centre of Candida albicans Interactions. Front. Cell. Infect. Microbiol. 2018, 8, 185. [Google Scholar] [CrossRef] [PubMed]
- Misslinger, M.; Hortschansky, P.; Brakhage, A.A.; Haas, H. Fungal Iron Homeostasis with a Focus on Aspergillus fumigatus. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2021, 1868, 118885. [Google Scholar] [CrossRef]
- Horianopoulos, L.C.; Kronstad, J.W. Connecting Iron Regulation and Mitochondrial Function in Cryptococcus neoformans. Curr. Opin. Microbiol. 2019, 52, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Devaux, F.; Thiébaut, A. The Regulation of Iron Homeostasis in the Fungal Human Pathogen Candida glabrata. Microbiology 2019, 165, 1041–1060. [Google Scholar] [CrossRef]
- Gupta, M.; Outten, C.E. Iron–Sulfur Cluster Signaling: The Common Thread in Fungal Iron Regulation. Curr. Opin. Chem. Biol. 2020, 55, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Pijuan, J.; Moreno, D.F.; Yahya, G.; Moisa, M.; Ul Haq, I.; Krukiewicz, K.; Mosbah, R.; Metwally, K.; Cavalu, S. Regulatory and Pathogenic Mechanisms in Response to Iron Deficiency and Excess in Fungi. Microb. Biotechnol. 2023, 16, 2053–2071. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Chen, Y.; Feng, C.; Bao, J.; Li, X.; Jiang, H. Establishment and Application of Real-Time Fluorescence Quantitative PCR Detection Technology for Metschnikowia bicuspidata Disease in Eriocheir sinensis. J. Fungi 2023, 9, 791. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Soto, D.; Ruiz-Herrera, J. Functional Analysis of the MAPK Pathways in Fungi. Rev. Iberoam. Micol. 2017, 34, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Sellam, A.; Chaillot, J.; Mallick, J.; Tebbji, F.; Richard Albert, J.; Cook, M.A.; Tyers, M. A Systematic Cell Size Screen Uncovers Coupling of Growth to Division by the P38/HOG Network in Candida albicans. Cell Biol. 2016. [Google Scholar] [CrossRef]
- Miller, K.E.; Kang, P.J.; Park, H.-O. Regulation of Cdc42 for Polarized Growth in Budding Yeast. Microb. Cell 2020, 7, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhao, B.; Zhou, X.; Lu, D.; Wang, Y.; Chen, Y.; Xiao, D. Analysis of the Molecular Basis of Saccharomyces Cerevisiae Mutant with High Nucleic Acid Content by Comparative Transcriptomics. Food Res. Int. 2021, 142, 110188. [Google Scholar] [CrossRef]
- Pujol-Carrion, N.; Pavón-Vergés, M.; Arroyo, J.; De La Torre-Ruiz, M.A. The MAPK Slt2/Mpk1 Plays a Role in Iron Homeostasis through Direct Regulation of the Transcription Factor Aft1. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2021, 1868, 118974. [Google Scholar] [CrossRef]
- Kaba, H.E.; Nimtz, M.; Müller, P.P.; Bilitewski, U. Involvement of the Mitogen Activated Protein Kinase Hog1p in the Response of Candida albicans to Iron Availability. BMC Microbiol. 2013, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Puri, S.; Lai, W.K.M.; Rizzo, J.M.; Buck, M.J.; Edgerton, M. Iron-responsive Chromatin Remodelling and MAPK Signalling Enhance Adhesion in Candida albicans. Mol. Microbiol. 2014, 93, 291–305. [Google Scholar] [CrossRef]
- Tripathi, A.; Liverani, E.; Tsygankov, A.Y.; Puri, S. Iron Alters the Cell Wall Composition and Intracellular Lactate to Affect Candida albicans Susceptibility to Antifungals and Host Immune Response. J. Biol. Chem. 2020, 295, 10032–10044. [Google Scholar] [CrossRef]
- Lavín, J.L.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative Genomics of the Oxidative Phosphorylation System in Fungi. Fungal Genet. Biol. 2008, 45, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Philpott, C.C.; Leidgens, S.; Frey, A.G. Metabolic Remodeling in Iron-Deficient Fungi. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2012, 1823, 1509–1520. [Google Scholar] [CrossRef]
- Duval, C.; Macabiou, C.; Garcia, C.; Lesuisse, E.; Camadro, J.; Auchère, F. The Adaptive Response to Iron Involves Changes in Energetic Strategies in the Pathogen Candida albicans. Microbiologyopen 2020, 9, e970. [Google Scholar] [CrossRef]
- Shakoury-Elizeh, M.; Protchenko, O.; Berger, A.; Cox, J.; Gable, K.; Dunn, T.M.; Prinz, W.A.; Bard, M.; Philpott, C.C. Metabolic Response to Iron Deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 14823–14833. [Google Scholar] [CrossRef] [PubMed]
- Pollack, J.; Harris, S.; Marten, M. Autophagy in Filamentous Fungi. Fungal Genet. Biol. 2009, 46, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Richie, D.L.; Askew, D.S. Autophagy: A Role in Metal Ion Homeostasis? Autophagy 2008, 4, 115–117. [Google Scholar] [CrossRef][Green Version]
- Chen, L.; Zhang, X.; Wang, W.; Geng, X.; Shi, Y.; Na, R.; Dou, D.; Li, H. Network and Role Analysis of Autophagy in Phytophthora sojae. Sci. Rep. 2017, 7, 1879. [Google Scholar] [CrossRef] [PubMed]
- Richie, D.L.; Fuller, K.K.; Fortwendel, J.; Miley, M.D.; McCarthy, J.W.; Feldmesser, M.; Rhodes, J.C.; Askew, D.S. Unexpected Link between Metal Ion Deficiency and Autophagy in Aspergillus fumigatus. Eukaryot. Cell 2007, 6, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Nagi, M.; Tanabe, K.; Nakayama, H.; Ueno, K.; Yamagoe, S.; Umeyama, T.; Ohno, H.; Miyazaki, Y. Iron-Depletion Promotes Mitophagy to Maintain Mitochondrial Integrity in Pathogenic Yeast Candida glabrata. Autophagy 2016, 12, 1259–1271. [Google Scholar] [CrossRef] [PubMed]
- Saikia, S.; Oliveira, D.; Hu, G.; Kronstad, J. Role of Ferric Reductases in Iron Acquisition and Virulence in the Fungal Pathogen Cryptococcus neoformans. Infect. Immun. 2014, 82, 839–850. [Google Scholar] [CrossRef]
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in Fungal Virulence. FEMS Microbiol. Rev. 2018, 42, 1–21. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).