Dazl and dnd Identify Both Embryonic and Gonadal Germ Cells in Chinese Hook Snout Carp (Opsariichthys bidens)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish and Sampling
2.2. RNA Isolation and cDNA Synthesis
2.3. Molecular Cloning and Analysis of Obdazl and Obdnd Gene
2.4. RT-PCR
2.5. RNA Synthesis and In Situ Hybridization
2.6. Preparation of Chimeric mRNAs and Microinjection
2.7. Microscopy
3. Results
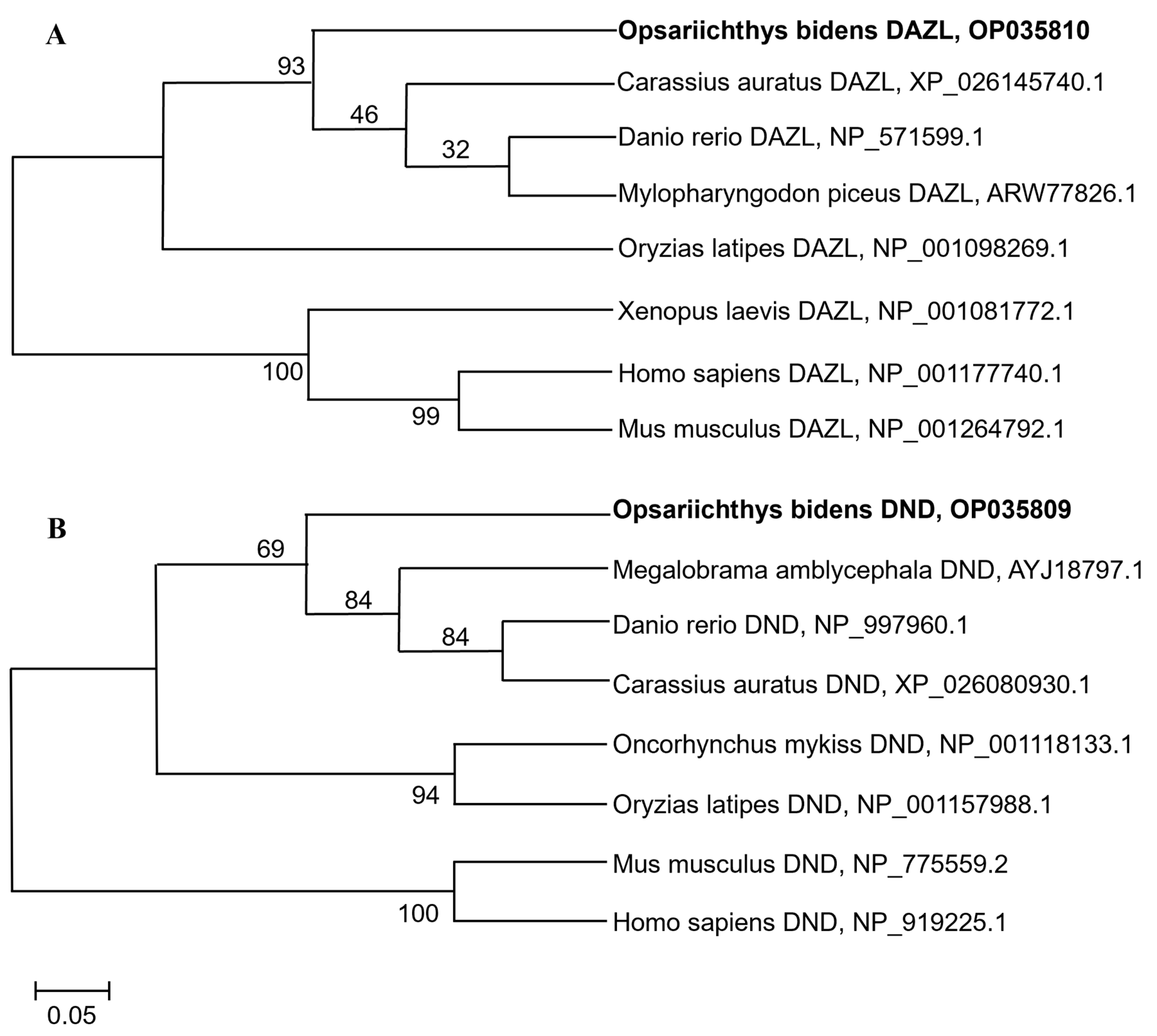
3.1. Cloning and Characterization of Obdazl and Obdnd
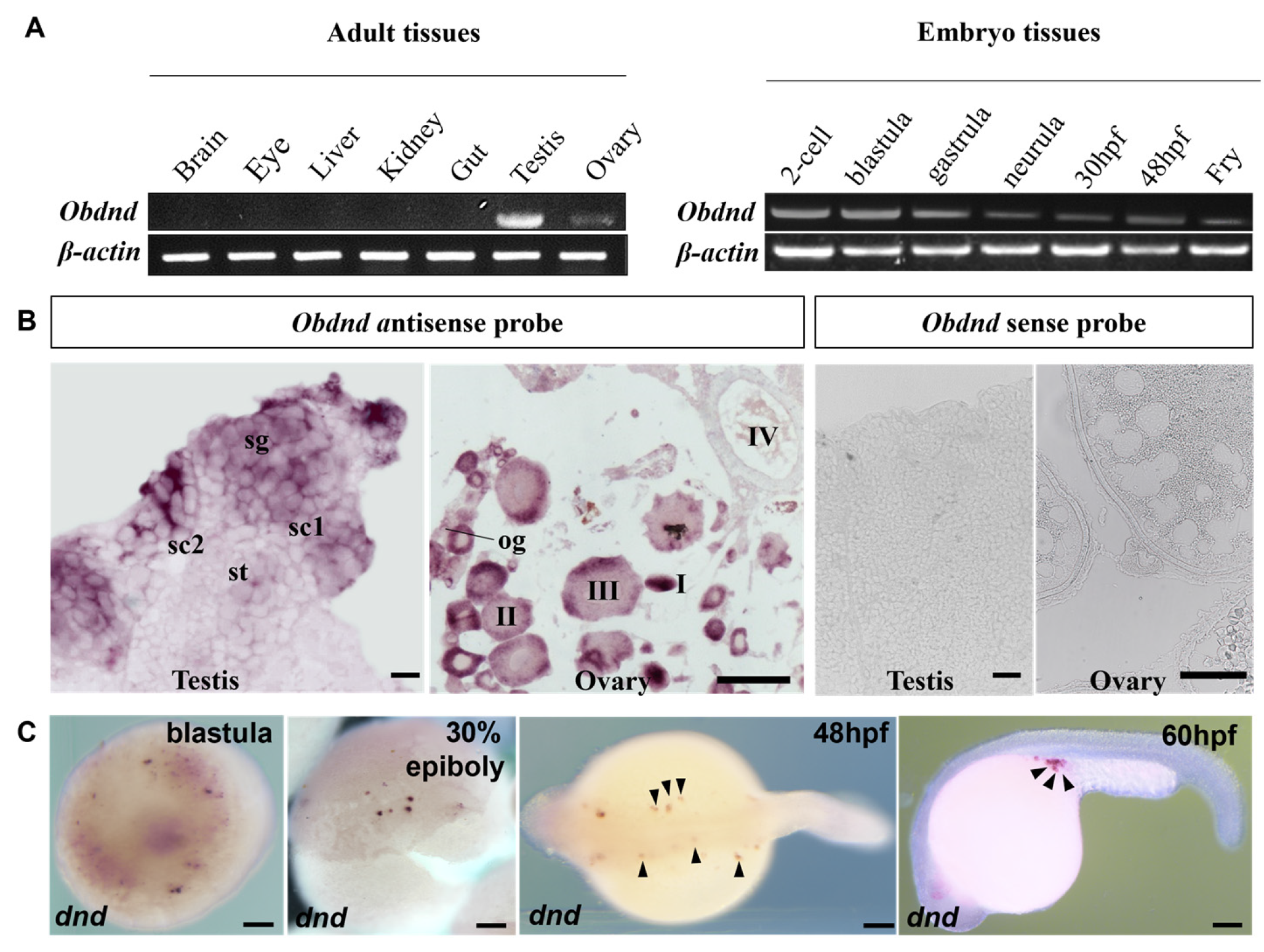
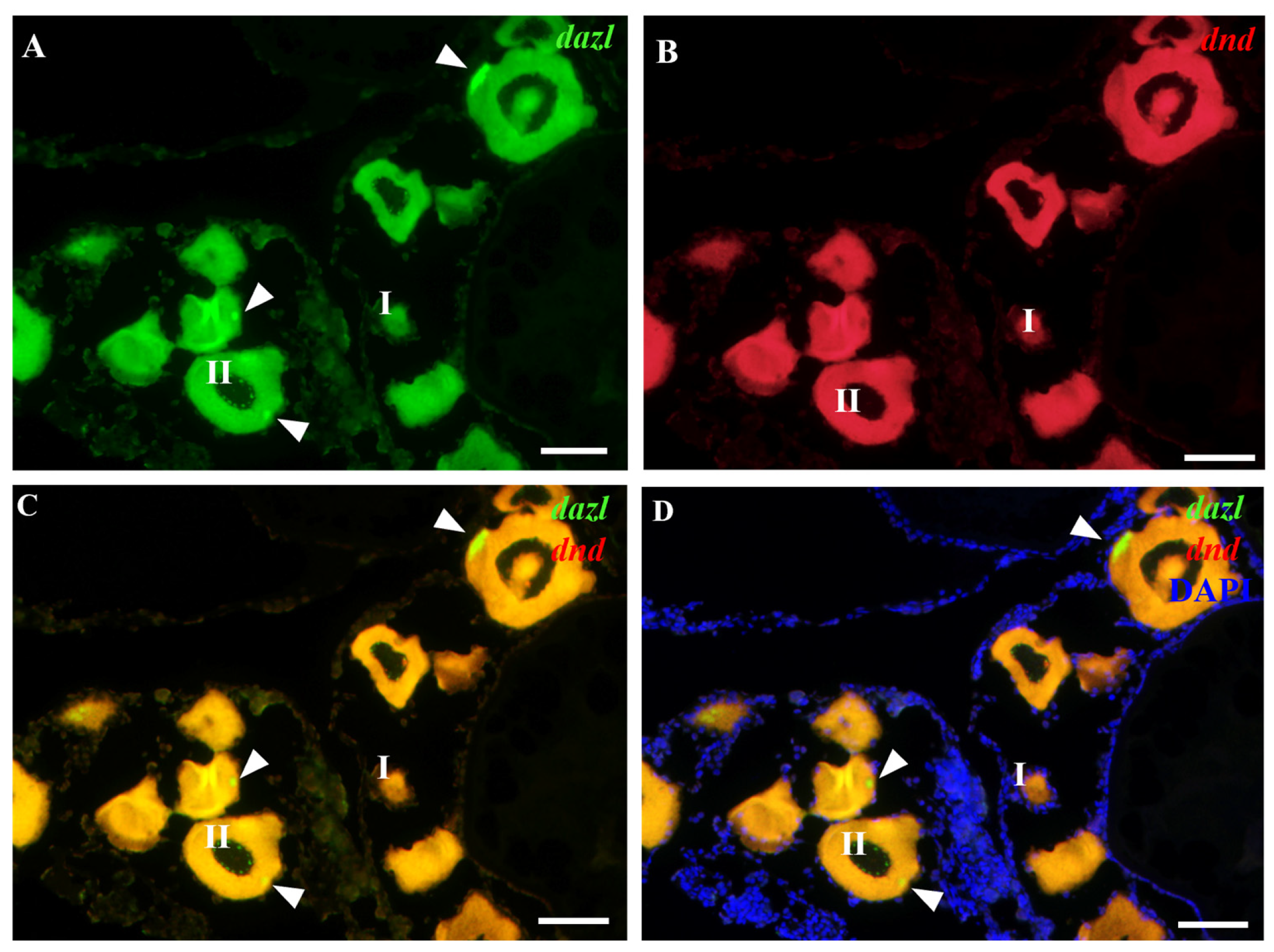
3.2. Germ Cell-Specific Expression of Obdazl and Obdnd RNAs
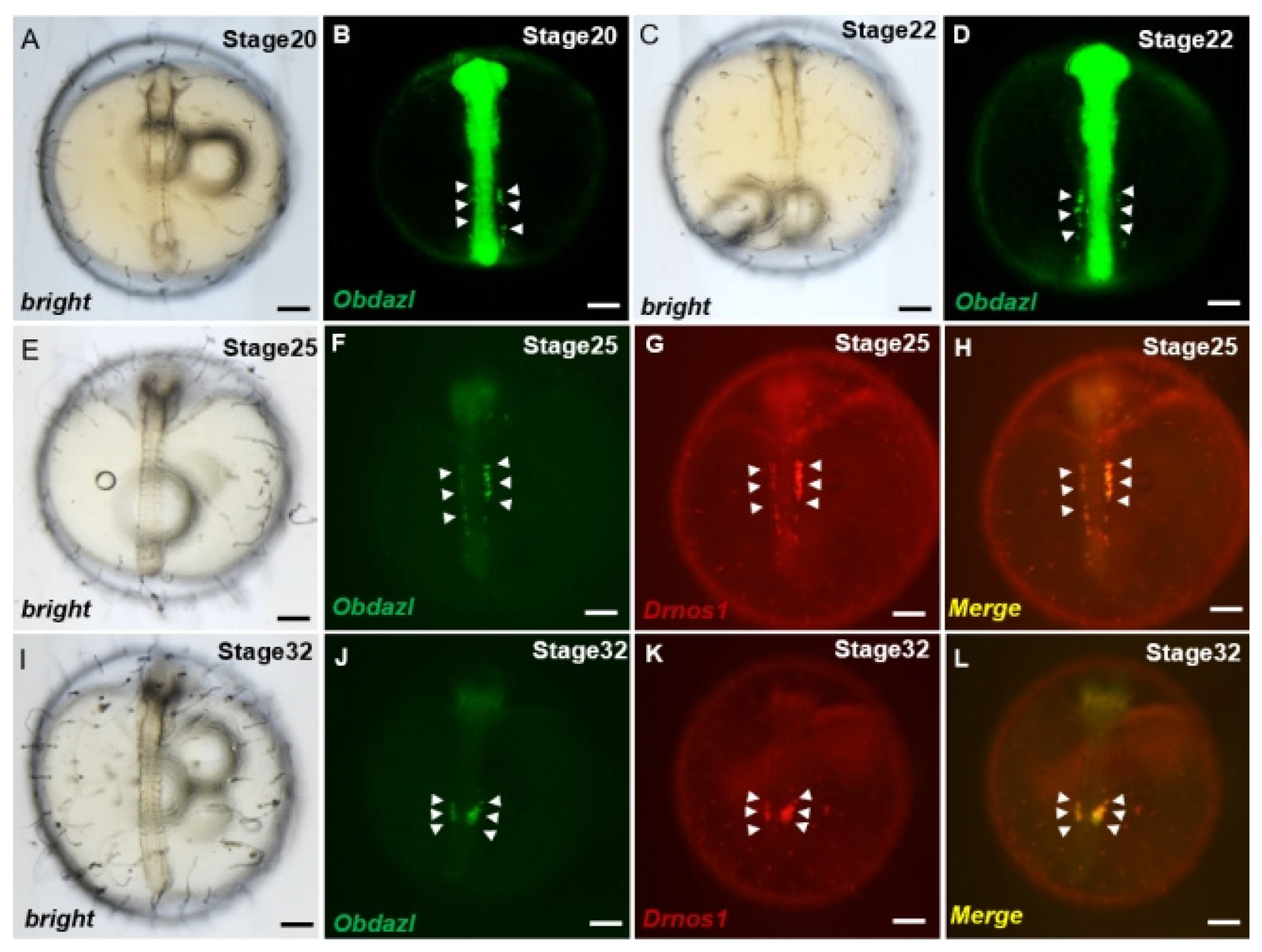
3.3. Obdazl 3′ UTR Enables GFP to Express in the PGCs of Medaka Stably
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wylie, C. Germ cells. Cell 1999, 96, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, T.S.; Quagio-Grassiotto, I. Presence of the matrix metalloproteinases during the migration of the primordial germ cells in zebrafish gonadal ridge. Cell Tissue Res. 2021, 383, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Baloch, A.R.; Franek, R.; Saito, T.; Psenicka, M. Dead-end (dnd) protein in fish-a review. Fish Physiol. Biochem. 2021, 47, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.F.; Cheng, S.F.; Wang, L.Q.; Yin, S.; De Felici, M.; Shen, W. DAZ Family Proteins, Key Players for Germ Cell Development. Int. J. Biol. Sci. 2015, 11, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Cooke, H.J.; Lee, M.; Kerr, S.; Ruggiu, M. A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum. Mol. Genet. 1996, 5, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Eberhart, C.G.; Maines, J.Z.; Wasserman, S.A. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature 1996, 381, 783–785. [Google Scholar] [CrossRef] [PubMed]
- Karashima, T.; Sugimoto, A.; Yamamoto, M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development 2000, 127, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.Y.; Moore, F.L.; Pera, R.A. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc. Natl. Acad. Sci. USA 2001, 98, 7414–7419. [Google Scholar] [CrossRef] [PubMed]
- Yen, P.H.; Chai, N.N.; Salido, E.C. The human autosomal gene DAZLA: Testis specificity and a candidate for male infertility. Hum. Mol. Genet. 1996, 5, 2013–2017. [Google Scholar] [CrossRef]
- Yen, P.H. Putative biological functions of the DAZ family. Int. J. Androl. 2004, 27, 125–129. [Google Scholar] [CrossRef]
- Wang, W.X.; Liang, S.S.; Zou, Y.X.; Li, Z.; Wu, Q.W.; Wang, L.J.; Wu, Z.H.; Peng, Z.Z.; You, F. Expression of scp3 and dazl reveals the meiotic characteristics of the olive flounder Paralichthys olivaceus(dagger). Biol. Reprod. 2023, 108, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhu, F.; Li, Z.; Hong, N.; Hong, Y. Dazl is a critical player for primordial germ cell formation in medaka. Sci. Rep. 2016, 6, 28317. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.R.; Rajkovic, G.; Daldello, E.M.; Luong, X.G.; Chen, J.; Conti, M. The RNA-binding protein DAZL functions as repressor and activator of mRNA translation during oocyte maturation. Nat. Commun. 2020, 11, 1399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, W.; Cui, Y.; Wen, L.; Yuan, Q.; Zhou, F.; Qiu, Q.; Sun, M.; Li, Z.; He, Z. Direct reprogramming of human Sertoli cells into male germline stem cells with the self-renewal and differentiation potentials via overexpressing DAZL/DAZ2/BOULE genes. Stem Cell Rep. 2021, 16, 2798–2812. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Deng, M.; Lv, W.; Wei, Z.; Cai, Y.; Cheng, P.; Wang, F.; Zhang, Y. Overexpression of bmp4, dazl, nanos3 and sycp2 in Hu Sheep Leydig Cells Using CRISPR/dcas9 System Promoted Male Germ Cell Related Gene Expression. Biology 2022, 11, 289. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, G.; Stebler, J.; Slanchev, K.; Dumstrei, K.; Wise, C.; Lovell-Badge, R.; Thisse, C.; Thisse, B.; Raz, E. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 2003, 13, 1429–1434. [Google Scholar] [CrossRef] [PubMed]
- Horvay, K.; Claussen, M.; Katzer, M.; Landgrebe, J.; Pieler, T. Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol. 2006, 291, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, C.; Aggarwal, S.; Zhu, R.; Kumar, M.; Zhao, M.; Meistrich, M.L.; Matin, A. The mouse dead-end gene isoform alpha is necessary for germ cell and embryonic viability. Biochem. Biophys. Res. Commun. 2007, 355, 194–199. [Google Scholar] [CrossRef]
- Aramaki, S.; Sato, F.; Kato, T.; Soh, T.; Kato, Y.; Hattori, M.A. Molecular cloning and expression of dead end homologue in chicken primordial germ cells. Cell Tissue Res. 2007, 330, 45–52. [Google Scholar] [CrossRef]
- Liu, L.; Hong, N.; Xu, H.; Li, M.; Yan, Y.; Purwanti, Y.; Yi, M.; Li, Z.; Wang, L.; Hong, Y. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells. Gene Expr. Patterns 2009, 9, 541–548. [Google Scholar] [CrossRef]
- Yang, X.; Yue, H.; Ye, H.; Li, C.; Wei, Q. Identification of a germ cell marker gene, the dead end homologue, in Chinese sturgeon Acipenser sinensis. Gene 2015, 558, 118–125. [Google Scholar] [CrossRef]
- Zhu, T.; Gui, L.; Zhu, Y.; Li, Y.; Li, M. Dnd is required for primordial germ cell specification in Oryzias Celebensis. Gene 2018, 679, 36–43. [Google Scholar] [CrossRef]
- Guralp, H.; Skaftnesmo, K.O.; Kjaerner-Semb, E.; Straume, A.H.; Kleppe, L.; Schulz, R.W.; Edvardsen, R.B.; Wargelius, A. Rescue of germ cells in dnd crispant embryos opens the possibility to produce inherited sterility in Atlantic salmon. Sci. Rep. 2020, 10, 18042. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Cho, Y.S.; Lee, H.B.; Park, J.Y.; Lim, H.K. Dead-End (dnd) Gene Cloning and Gonad-Specific Expression Pattern in Starry Flounder (Platichthys stellatus). Animals 2021, 11, 2256. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, Y.; Wen, Z.; Jawad, M.; Gui, L.; Li, M. Spinyhead Croaker Germ Cells Gene dnd Visualizes Primordial Germ Cells in Medaka. Life 2022, 12, 1226. [Google Scholar] [CrossRef] [PubMed]
- Hong, N.; Li, M.; Yuan, Y.; Wang, T.; Yi, M.; Xu, H.; Zeng, H.; Song, J.; Hong, Y. Dnd Is a Critical Specifier of Primordial Germ Cells in the Medaka Fish. Stem Cell Rep. 2016, 6, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Gross-Thebing, T.; Yigit, S.; Pfeiffer, J.; Reichman-Fried, M.; Bandemer, J.; Ruckert, C.; Rathmer, C.; Goudarzi, M.; Stehling, M.; Tarbashevich, K.; et al. The Vertebrate Protein Dead End Maintains Primordial Germ Cell Fate by Inhibiting Somatic Differentiation. Dev. Cell 2017, 43, 704–715.e5. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, R.; Katayama, N.; Sadaie, S.; Miwa, M.; Sanchez Matias, G.A.; Ichida, K.; Fujii, W.; Naito, K.; Hayashi, M.; Yoshizaki, G. Production of Germ Cell-Less Rainbow Trout by dead end Gene Knockout and their Use as Recipients for Germ Cell Transplantation. Mar. Biotechnol. 2022, 24, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hong, N.; Xu, H.; Song, J.; Hong, Y. Germline replacement by blastula cell transplantation in the fish medaka. Sci. Rep. 2016, 6, 29658. [Google Scholar] [CrossRef]
- Hou, M.; Feng, K.; Luo, H.; Jiang, Y.; Xu, W.; Li, Y.; Song, Y.; Chen, J.; Tao, B.; Zhu, Z.; et al. Complete Depletion of Primordial Germ Cells Results in Masculinization of Monopterus albus, a Protogynous Hermaphroditic Fish. Mar. Biotechnol. 2022, 24, 320–334. [Google Scholar] [CrossRef]
- Westerich, K.J.; Tarbashevich, K.; Schick, J.; Gupta, A.; Zhu, M.; Hull, K.; Romo, D.; Zeuschner, D.; Goudarzi, M.; Gross-Thebing, T.; et al. Spatial organization and function of RNA molecules within phase-separated condensates in zebrafish are controlled by Dnd1. Dev. Cell 2023, 58, 1578–1592.e5. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; He, S.; Mayden, R.L. The complete mitochondrial genome of the Chinese hook snout carp Opsariichthys bidens (Actinopterygii: Cypriniformes) and an alternative pattern of mitogenomic evolution in vertebrate. Gene 2007, 399, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Schmidt, B.V.; He, S. The potential colonization histories of Opsariichthys bidens (Cyprinidae) in China using Bayesian binary MCMC analysis. Gene 2018, 676, 1–8. [Google Scholar] [CrossRef]
- Fu, S.J.; Peng, Z.; Cao, Z.D.; Peng, J.L.; He, X.K.; Xu, D.; Zhang, A.J. Habitat-specific locomotor variation among Chinese hook snout carp (Opsariichthys bidens) along a river. PLoS ONE 2012, 7, e40791. [Google Scholar] [CrossRef]
- Perdices, A.; Sayanda, D.; Coelho, M.M. Mitochondrial diversity of Opsariichthys bidens (Teleostei, Cyprinidae) in three Chinese drainages. Mol. Phylogenet. Evol. 2005, 37, 920–927. [Google Scholar] [CrossRef]
- Tang, R.; Zhu, Y.; Gan, W.; Zhang, Y.; Yao, Z.; Ren, J.; Li, M. De novo transcriptome analysis of gonads reveals the sex-associated genes in Chinese hook snout carp Opsariichthys bidens. Aquac. Rep. 2022, 23, 101068. [Google Scholar] [CrossRef]
- Xu, X.; Guan, W.; Niu, B.; Guo, D.; Xie, Q.P.; Zhan, W.; Yi, S.; Lou, B. Chromosome-Level Assembly of the Chinese Hooksnout Carp (Opsariichthys bidens) Genome Using PacBio Sequencing and Hi-C Technology. Front. Genet. 2021, 12, 788547. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kan, Y.T.; Zhong, Y.; Jawad, M.; Wei, W.B.; Gu, K.Y.; Gui, L.; Li, M.Y. Generation of a normal long-term cultured Chinese hook snout carp spermatogonial stem cell line capable of sperm production in vitro. Biology 2022, 11, 1069. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Xu, C.; Zhu, Y.; Yan, J.; Yao, Z.; Zhou, W.; Gui, L.; Li, M. Identification and expression analysis of sex biased miRNAs in Chinese hook snout carp Opsariichthys bidens. Front. Genet. 2022, 13, 990683. [Google Scholar] [CrossRef]
- Li, M.; Hong, N.; Gui, J.; Hong, Y. Medaka piwi is essential for primordial germ cell migration. Curr. Mol. Med. 2012, 12, 1040–1049. [Google Scholar] [CrossRef]
- Yuan, C.; Chen, K.; Zhu, Y.; Yuan, Y.; Li, M. Medaka igf1 identifies somatic cells and meiotic germ cells of both sexes. Gene 2018, 642, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, W.; Zhu, Y.F.; Zhu, T.Y.; Ma, L.B.; Li, M.Y. Evolutionarily conserved vasa identifies embryonic and gonadal germ cells in spinyhead croaker Collichthys lucidus. J. Fish Biol. 2019, 94, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhu, Y.; Yuan, C.; Zhao, Y.; Zhou, W.; Li, M. Differential Expression of Duplicate Insulin-like Growth Factor-1 Receptors (igf1rs) in Medaka Gonads. Life 2022, 12, 859. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Fujimoto, T.; Maegawa, S.; Inoue, K.; Tanaka, M.; Arai, K.; Yamaha, E. Visualization of primordial germ cells in vivo using GFP-nos1 3′UTR mRNA. Int. J. Dev. Biol. 2006, 50, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, M.; Gui, J.; Hong, Y. Cloning and expression of medaka dazl during embryogenesis and gametogenesis. Gene Expr. Patterns 2007, 7, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Z.; Liu, W.; Li, Z.; Wang, Y.; Zhou, L.; Yi, M.S.; Gui, J.F. Molecular characterization and expression pattern of a germ cell marker gene dnd in gibel carp (Carassius gibelio). Gene 2016, 591, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, P.; Xia, J.; Guo, J.; Shi, Y.; Zhong, Y.; Li, M. Evolutionarily conserved boule and dazl identify germ cells of Coilia nasus. Aquac. Fish. 2021, 8, 244–251. [Google Scholar] [CrossRef]
- Slanchev, K.; Stebler, J.; Goudarzi, M.; Cojocaru, V.; Weidinger, G.; Raz, E. Control of Dead end localization and activity—Implications for the function of the protein in antagonizing miRNA function. Mech. Dev. 2009, 126, 270–277. [Google Scholar] [CrossRef]
- Li, M.; Shen, Q.; Xu, H.; Wong, F.M.; Cui, J.; Li, Z.; Hong, N.; Wang, L.; Zhao, H.; Ma, B.; et al. Differential conservation and divergence of fertility genes boule and dazl in the rainbow trout. PLoS ONE 2011, 6, e15910. [Google Scholar] [CrossRef]
- Lin, F.; Zhao, C.Y.; Xu, S.H.; Ma, D.Y.; Xiao, Z.Z.; Xiao, Y.S.; Xu, C.A.; Liu, Q.H.; Li, J. Germline-specific and sexually dimorphic expression of a dead end gene homologue in turbot (Scophthalmus maximus). Theriogenology 2013, 80, 665–672. [Google Scholar] [CrossRef]
- Bhat, N.; Hong, Y. Cloning and expression of boule and dazl in the Nile tilapia (Oreochromis niloticus). Gene 2014, 540, 140–145. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, Y.; Ye, H.; Lu, L.; Li, H.; Xu, Z.; Li, W.; Yin, X.; Xu, D. Identification of germ cells in large yellow croaker (Larimichthys crocea) and yellow drum (Nibea albiflora) using RT-PCR and in situ hybridization analyses. Gene 2023, 863, 147280. [Google Scholar] [CrossRef] [PubMed]
- Jamieson-Lucy, A.; Mullins, M.C. The vertebrate Balbiani body, germ plasm, and oocyte polarity. Curr. Top. Dev. Biol. 2019, 135, 1–34. [Google Scholar] [PubMed]
- Peng, J.X.; Xie, J.L.; Zhou, L.; Hong, Y.H.; Gui, J.F. Evolutionary conservation of Dazl genomic organization and its continuous and dynamic distribution throughout germline development in gynogenetic gibel carp. J. Exp. Zool. B Mol. Dev. Evol. 2009, 312, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Dwarakanath, M.; Lim, M.; Xu, H.; Hong, Y. Differential expression of boule and dazl in adult germ cells of the Asian seabass. Gene 2014, 549, 237–242. [Google Scholar] [CrossRef]
- Koprunner, M.; Thisse, C.; Thisse, B.; Raz, E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001, 15, 2877–2885. [Google Scholar] [CrossRef]

| Primer | Sequence (5′ to 3′ Direction) | Purpose |
|---|---|---|
| Obdazl-5′-R1 | GGCAAGATGACGCCCAACACACT | 5′ RACE |
| Obdazl-5′-NR1 | ACTCAAACTGGGACCTGCCATCAT | |
| Obdazl-3′-F1 | GTGAAAATCATCACTTATCGCGTT | 3′ RACE |
| Obdazl-3′-NF1 | CCTGCAGGATTTATGGTCCCACCGAT | |
| Obdazl full F | ATGTATCAGGGCGTTAAGTTACC | CDS cloning |
| Obdazl full R | GACTCTGTTAACTCTTCTGTAG | |
| Obdazl 3′ UTR F | ctcgagTAGCTAACCCTTCATCCTTTTTTT | mRNA |
| Obdazl 3′ UTR R | ggtacctctagaGCAGTTAAAAGGTGTAAATA | |
| Obdnd 5′ R1 | ATGGCGTGCGCTCAGACCGGC | 5′ RACE |
| Obdnd 5′ NR1 | ATCCTGCAGCAGCTGATGACAG | |
| Obdnd 3′ F1 | CGAAGCCCATGTCCAACATGCG | 3′ RACE |
| Obdnd 3′ NF1 | CGTCCAGGACCAAGTGGCGAAC | |
| Obdnd full F | ATGGACGATTGGGAGGAGG | CDS cloning |
| Obdnd full R | CTCCCAGTTCTCCTCCTCGTCAG | |
| β–actin F | TTCAACAGCCCTGCCATGTA | Internal control |
| β–actin R | CCTCCAATCCAGACAGTAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Z.; Xue, L.; Song, P.; Jawad, M.; Xu, C.; Bu, W.; Li, M. Dazl and dnd Identify Both Embryonic and Gonadal Germ Cells in Chinese Hook Snout Carp (Opsariichthys bidens). Fishes 2024, 9, 214. https://doi.org/10.3390/fishes9060214
Yin Z, Xue L, Song P, Jawad M, Xu C, Bu W, Li M. Dazl and dnd Identify Both Embryonic and Gonadal Germ Cells in Chinese Hook Snout Carp (Opsariichthys bidens). Fishes. 2024; 9(6):214. https://doi.org/10.3390/fishes9060214
Chicago/Turabian StyleYin, Zifeng, Lingzhan Xue, Peng Song, Muhammad Jawad, Cong Xu, Weishao Bu, and Mingyou Li. 2024. "Dazl and dnd Identify Both Embryonic and Gonadal Germ Cells in Chinese Hook Snout Carp (Opsariichthys bidens)" Fishes 9, no. 6: 214. https://doi.org/10.3390/fishes9060214
APA StyleYin, Z., Xue, L., Song, P., Jawad, M., Xu, C., Bu, W., & Li, M. (2024). Dazl and dnd Identify Both Embryonic and Gonadal Germ Cells in Chinese Hook Snout Carp (Opsariichthys bidens). Fishes, 9(6), 214. https://doi.org/10.3390/fishes9060214






