Genetic Characterization of Procambarus clarkii Cultured in Sichuan Province Based on Microsatellite Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Microsatellite Markers
2.3. Data Analysis
3. Results
3.1. Genetic Diversity Analysis of Different P. clarkii Populations
3.2. AMOVA and Genetic Differenation of P. clarkii Populations
3.3. Genetic Structure of P. clarkii Populations
3.4. Genetic Status of Cultured P. clarkii Populations in Sichuan
4. Discussion
4.1. Genetic Diversity of Cultured Populations of P. clarkii
4.2. Genetic Differentiation and Historical Dynamics of P. clarkii Population
4.3. Recommendations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oficialdegui, F.J.; Clavero, M.; Sánchez, M.I.; Green, A.J.; Boyero, L.; Michot, T.C.; Klose, K.; Kawai, T.; Lejeusne, C. Unravelling the global invasion routes of a worldwide invader, the red swamp crayfish (Procambarus clarkii). Freshw. Biol. 2019, 64, 1382–1400. [Google Scholar] [CrossRef]
- Pennak, R.W. Fresh-water Invertebrates of the United States. J. Aquat. Ecosyst. Stress Recovery (Former. J. Aquat. Ecosyst. Health) 1955, 7, 126. [Google Scholar] [CrossRef]
- Tian, J.; Xu, Q.Q.; Tian, L.; Hu, W.; Yang, C.G.; Gao, W.H. The muscle composition analysis and flesh quality of Procambarus clarkii in the Dongting Lake. Acta Hydrobiol. Sin. 2017, 41, 870–877. [Google Scholar]
- Xiao, F.; Liu, Z.S.; Guo, Y.F.; Zhu, Z.W.; Ma, D.W.; Qing, T.J. Report on the development of crayfish industry in China (2017). China Fish. 2017, 7, 8–17. [Google Scholar]
- Luo, Y.T.; Liu, Y.; Liu, J.M.; Xiao, T.Y.; Lin, L.Q. Analysis of Muscle Nutritional Components of Different Sizes of Procambarus clarkii Cultured in Rice Field in Hunan Province. Chin. J. Anim. Nutr. 2021, 33, 4592–4603. [Google Scholar]
- Yu, X.J.; Hao, X.J.; Yang, L.K.; Dang, Z.Q.; Wang, X.G.; Zhang, Y.H.; Xi, Z. Report on the development of crayfish industry in China (2023). China Fish. 2023, 7, 26–31. [Google Scholar]
- Peng, W.F.; Zhang, D.M.; Luo, Y.M.; Tao, S.; Liang, X.X. Influence of natural factors on vegetation NDVI using geographical detection in Sichuan Province. Acta Geogr. Sin. 2019, 74, 1758–1776. [Google Scholar]
- Alvanou, M.V.; Apostolidis, A.P.; Lattos, A.; Michaelidis, B.; Giantsis, I.A. The Coding Mitogenome of the Freshwater Crayfish Pontastacus leptodactylus (Decapoda:Astacidea:Astacidae) from Lake Vegoritida, Greece and Its Taxonomic Classification. Genes 2023, 14, 494. [Google Scholar] [CrossRef]
- Ren, S.; Mather, P.B.; Tang, B.; Hurwood, D.A. Levels of genetic diversity and inferred origins of Penaeus vannamei culture resources in China: Implications for the production of a broad synthetic base population for genetic improvement. Aquaculture 2018, 491, 221–231. [Google Scholar] [CrossRef]
- Zhou, L.; Gao, J.C.; Yang, Y.P.; Nie, Z.J.; Liu, K.; Xu, G.C. Genetic Diversity and Population Structure Analysis of Chinese Mitten Crab (Eriocheir sinensis) in the Yangtze and Liaohe Rivers. Fishes 2023, 8, 253. [Google Scholar] [CrossRef]
- Weber, J.L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics 1990, 7, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Han, C.H.; Ma, H.T.; Jiang, H.B.; Liu, Y.; Han, H.Z.; Wang, F. Isolation of microsatellite markers for black rockfish Sebastes schlegeli and genetic diversity of wild and cultured populations. Oceanol. Limnol. Sin. 2016, 47, 213–220. [Google Scholar]
- Fazzi-Gomes, P.F.; Aguiar, J.; Marques, D.; Fonseca, C.G.; Moreira, F.C.; Rodrigues, M.; Silva, C.S.; Hamoy, I.; Santos, S. Novel Microsatellite Markers Used for Determining Genetic Diversity and Tracing of Wild and Farmed Populations of the Amazonian Giant Fish Arapaima gigas. Genes 2021, 12, 1324. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.R.; Wang, T.; Song, L.L.; Yang, Y.; Zhong, C.Y.; Tao, Y.H.; Liu, X.C. Genetic diversity among breeding populations of giant grouper (Epinephelus lanceolatus). Fish. China 2024, 48, 49616. [Google Scholar]
- Zhou, Y.; Tong, J.T.; Wang, J.R.; Yu, X.M. Development of microsatellite markers and genetic diversity in wild and cultured populations of black carp (Mylopharyngodon piceus) along the Yangtze River. Aquac. Int. 2020, 28, 1867–1882. [Google Scholar] [CrossRef]
- Cui, W.T.; Zhou, Y.F.; Bai, Z.Y.; Wang, Z.Y.; Zhong, L.D. Genetic diversity and structure analysis of Procambarus clarkii populations in Anhui Province. J. Fish. China 2023, 47, 90–99. [Google Scholar]
- Duan, B.H.; Kang, T.X.; Wan, H.F.; Liu, W.B.; Zhang, F.H.; Mu, S.M.; Guan, Y.Q.; Li, Z.J.; Tian, Y.; Kang, X.J. Microsatellite markers reveal genetic diversity and population structure of Portunus trituberculatus in the Bohai Sea, China. Sci. Rep. 2023, 13, 8668. [Google Scholar] [CrossRef]
- Liu, J.Q.; Sun, Y.F.; Chen, Q.Q.; Wang, M.; Li, Q.; Zhou, W.Z.; Cheng, Y.X. Genetic Diversity Analysis of the Red Swamp Crayfish Procambarus clarkii in Three Cultured Populations Based on Microsatellite Markers. Animals 2023, 13, 1881. [Google Scholar] [CrossRef]
- Wang, W.; Ma, C.Y.; Ouyang, L.L.; Chen, W.; Zhao, M.; Zhang, F.; Fu, Y.; Jiang, K.J.; Liu, Z.Q.; Zhang, H.; et al. Genetic diversity and population structure analysis of Lateolabrax maculatus from Chinese coastal waters using polymorphic microsatellite markers. Sci. Rep. 2021, 11, 15260. [Google Scholar] [CrossRef]
- Zhong, Y.Z.; Tang, Z.S.; Huang, L.M.; Wang, D.P.; Lu, Z.L. Genetic diversity of Procambarus clarkii populations based on mitochondrial DNA and microsatellite markers in different areas of Guangxi, China. Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2020, 31, 48–56. [Google Scholar] [CrossRef]
- Wang, Q.S.; Hu, Q.; Yang, S.Q.; Li, Y.H. Isolation of tetrameric microsatellite markers and its application on parentage identification in Procambarus clarkii. Aquac. Int. 2023, 31, 2099–2111. [Google Scholar] [CrossRef]
- Belfiore, N.M.; May, B. Variable microsatellite loci in red swamp crayfish, Procambarus clarkii, and their characterization in other crayfish taxa. Mol. Ecol. 2000, 9, 2231–2234. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.F.; Liu, M.; Zhou, Y.L.; Wen, W.Y.; Li, Z.; Li, Z.; Wang, Z.W.; Gui, J.F. Genetic Diversity Evaluation and Population Structure Analysis of Red Swamp Crayfish (Procambarus clarkii) from Lakes and Rice Fields by SSR Markers. Fishes 2022, 4, 142. [Google Scholar] [CrossRef]
- Oosterhout, C.V.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T. POPGENE Version 1.32 Microsoft Windows-Based Freeware for Populations Genetic Analysis; University of Alberta: Edmonton, Canada, 1999. [Google Scholar]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices, Version 2.9.3; Updated from Goudet (1995). 2001. Available online: http://www.unil.ch/izea/softwares/fstat.html (accessed on 23 August 2024).
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Porras-Hurtado, L.; Ruiz, Y.; Santos, C.; Phillips, C.; Carracedo, A.; Lareu, M.V. An overview of STRUCTURE: Applications, parameter settings, and supporting software. Front. Genet. 2013, 4, 98. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J. Computer note. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Freeland, J.R.; Petersen, S.D.; Kirk, H. Molecular Ecology, 2nd ed.; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar]
- Liu, F.; Qu, Y.K.; Geng, C.; Wang, A.M.; Zhang, J.H.; Li, J.F.; Chen, K.J.; Liu, B.; Tian, H.Y.; Yang, W.P.; et al. Analysis of the population structure and genetic diversity of the red swamp crayfish (Procambarus clarkii) in China using SSR markers. Electron. J. Biotechnol. 2020, 47, 59–71. [Google Scholar] [CrossRef]
- Shen, Y.B.; Wang, L.; Fu, J.J.; Xu, X.Y.; Yue, G.H.; Li, J.L. Population structure, demographic history and local adaptation of the grass carp. BMC Genom. 2019, 20, 467. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Mu, K.M.; Ni, Z.X.; Liu, X.H.; Li, Y.G.; Xu, L.A. Analysis of genetic diversity of ancient Ginkgo populations using SSR markers. Ind. Crops Prod. 2020, 145, 111942. [Google Scholar] [CrossRef]
- Shete, S.; Tiwari, H.; Elston, R.C. On Estimating the Heterozygosity and Polymorphism Information Content Value. Theor. Popul. Biol. 2000, 57, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; Gaggiotti, O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 2006, 15, 1419–1439. [Google Scholar] [CrossRef]
- Duan, B.H.; Mu, S.M.; Guan, Y.Q.; Li, S.Q.; Yu, Y.; Liu, W.B.; Li, Z.J.; Ji, X.D.; Kang, X.J. Genetic diversity and population structure of the swimming crab (Portunus trituberculatus) in China seas determined by genotyping-by-sequencing (GBS). Aquaculture 2022, 555, 738233. [Google Scholar] [CrossRef]
- Tan, Y.F.; Peng, G.H.; Xiong, L.J.; Peng, B.; Wu, Y.B.; Song, C.W.; Feng, B.X. Genetic diversity and structure analysis of 13 red swamp crayfish (Procambarus clarkii) populations in Yangtze River basin. J. Huazhong Agric. Univ. 2020, 39, 33–39. [Google Scholar]
- Sun, J.X.; Peng, G.H.; Xiong, L.J.; Chen, C.; Li, Y.H.; Bai, X.F. Genome-wide SSR marker development and application in genetic diversity analysis of the red swamp crayfish, Procambarus clarkii (Girard, 1852) in China. Crustaceana 2021, 94, 189–205. [Google Scholar] [CrossRef]
- Liu, X.Y.; Xiong, L.J.; Peng, B.; Luo, C.R.; Peng, G.H.; Zhu, X.; Bai, X.F. Genetic Diversity Analysis of 14 Cultured Populations of Red Swamp Crayfish Procambarus clarkii in China. Fish. Sci. 2023, 42, 457–465. [Google Scholar]
- Herborg, L.; Weetman, D.; Oosterhout, C.V.; Hänfling, B. Genetic population structure and contemporary dispersal patterns of a recent European invader, the Chinese mitten crab, Eriocheir sinensis. Mol. Ecol. 2007, 2, 231–242. [Google Scholar] [CrossRef]
- Balloux, F.; Lugon-Moulin, N. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 2002, 11, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; McInnes, K.H. Introduction to Conservation Genetics; Cambridge University Press: New York, NY, USA, 2002. [Google Scholar]
- Zhang, J.Q.; Xu, C.X.; Wang, S.F.; Wang, S.Q.; Li, Y.M. Variations in Genetic Diversity of Invasive Species Lithobates catesbeianus in China. Animals 2024, 14, 1287. [Google Scholar] [CrossRef] [PubMed]
- Berg, D.J.; Garton, D.W.; Macisaac, H.J.; Panov, V.E.; Telesh, I.V. Changes in genetic structure of North American Bythotrephes populations following invasion from Lake Ladoga, Russia. Freshw. Biol. 2002, 47, 275–282. [Google Scholar] [CrossRef]
- Kolbe, J.J.; Glor, R.E.; Rodríguez Schettino, L.; Lara, A.C.; Larson, A.; Losos, J.B. Genetic variation increases during biological invasion by a Cuban lizard. Nature 2004, 431, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, Y.J.; Wu, Y.Y.; Zhou, Y.; Lv, F.; Wu, H.Y.; Li, J.M.; Song, A.H. Genetic Diversity Analysis of 3 Populations and Their Hybrid Progenies inBlood Clam Scapharca broughtonii. Fish. Sci. 2024, 43, 561–570. [Google Scholar]
- Wang, P.P.; Ding, Y.D.; Yu, X.D.; Jia, X.Q.; Zhang, G.S.; Yin, S.W.; Chen, S.Q.; Zhou, G.Q. Growth Trait and Genetic Diversity in the F Progeny of Dark SleeperOdontobutis potamophila from Three Different Populations. Fish. Sci. 2016, 35, 528–534. [Google Scholar]
- Shi, G.R.; Ma, C.C. Biological characteristics of alien plants successful invasion. Chin. J. Appl. Ecol. 2006, 17, 727–732. [Google Scholar]
- Kouba, A.; Liptak, B.; Kubec, J.; Blaha, M.; Vesely, L.; Haubrock, P.J.; Oficialdegui, F.J.; Niksirat, H.; Patoka, J.; Buric, M. Survival, Growth, and Reproduction: Comparison of Marbled Crayfish with Four Prominent Crayfish Invaders. Biology 2021, 10, 422. [Google Scholar] [CrossRef]
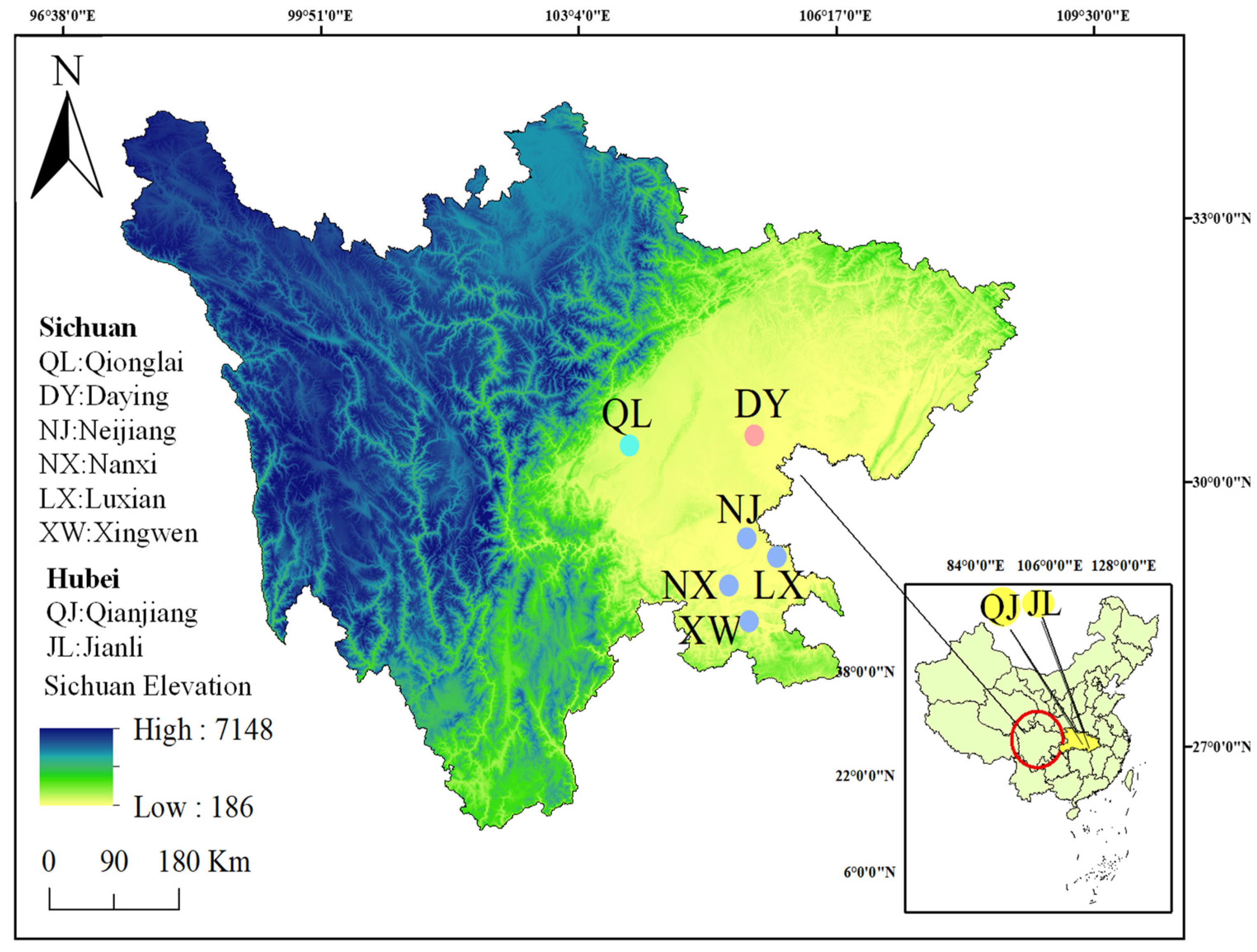
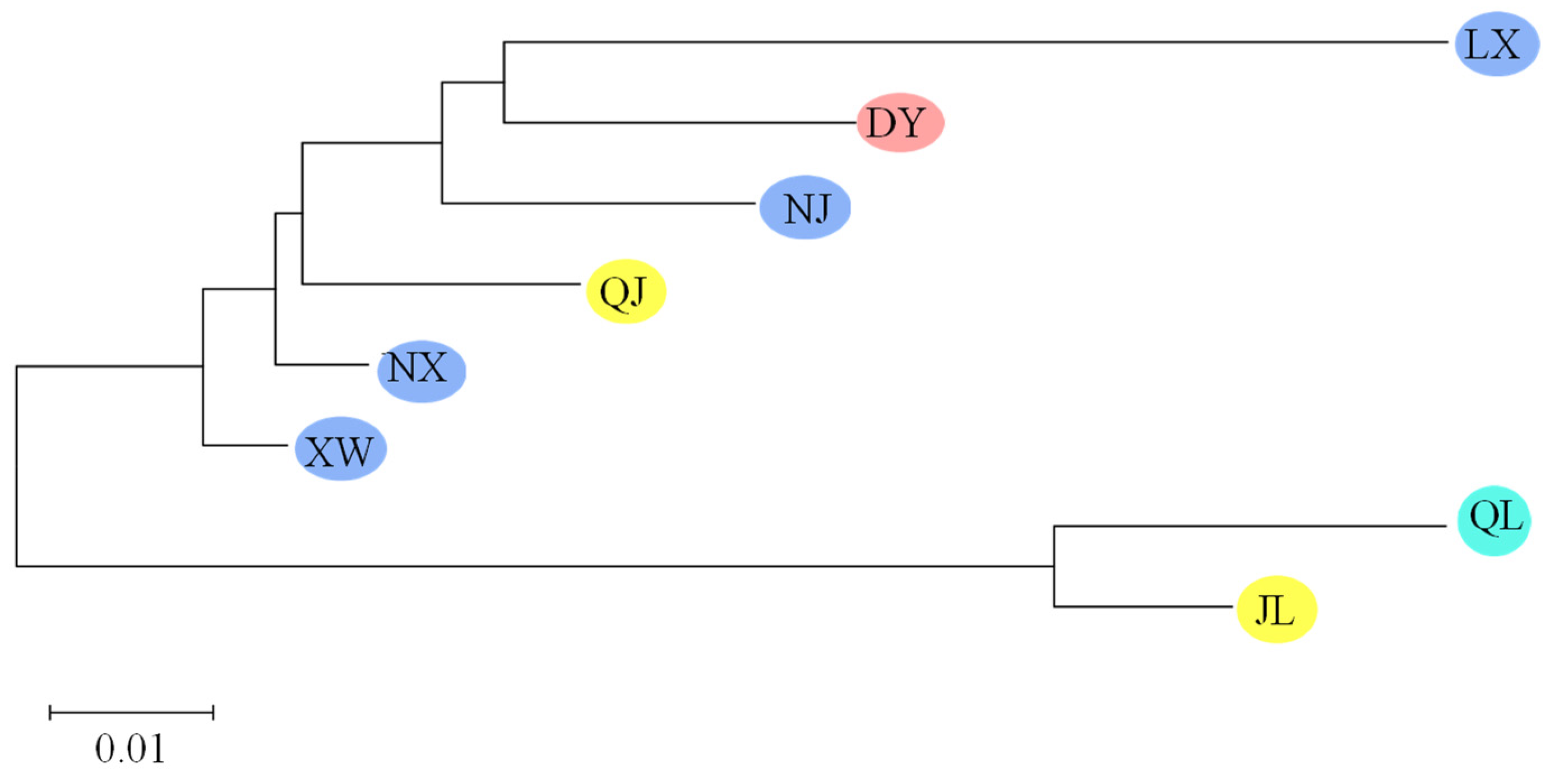
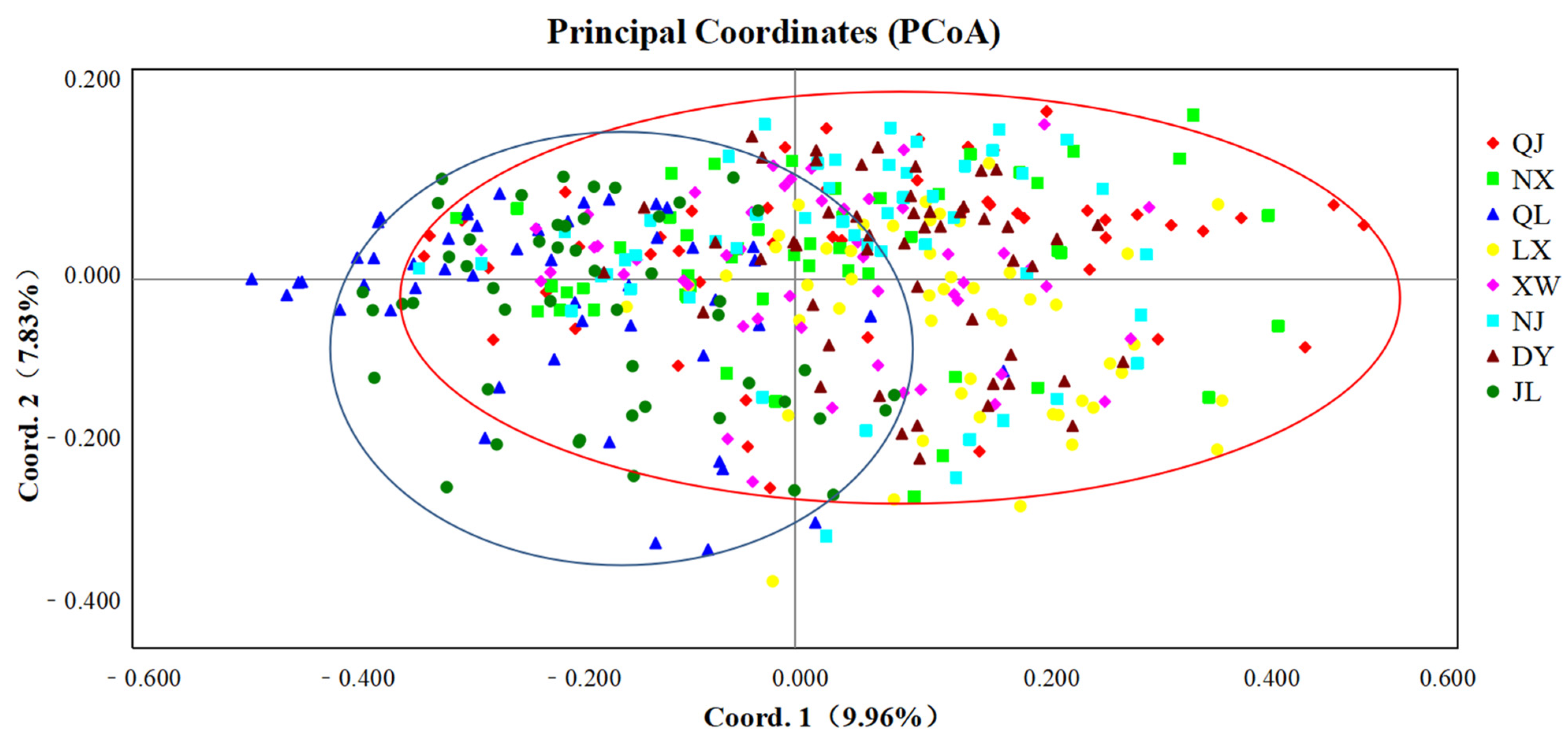

| Code | Location | Sample Size | Date of Sampling | Total Length (mm) | Weight (g) |
|---|---|---|---|---|---|
| QJ * | Hubei Qianjiang | 50 | 13 April 2023 | 108.3 ± 1.0 | 41.9 ± 14.7 |
| JL * | Hubei Jianli | 50 | 18 July 2023 | 97.0 ± 7.7 | 29.9 ± 6.6 |
| NX | Sichuan Nanxi | 50 | 13 April 2023 | 91.4 ± 4.0 | 27.0 ± 3.2 |
| QL | Sichuan Qionglai | 50 | 1 April 2023 | 95.8 ± 7.0 | 27.1 ± 7.3 |
| LX | Sichuan Luxian | 50 | 12 May 2023 | 102.8 ± 5.9 | 35.9 ± 6.8 |
| XW | Sichuan Xingwen | 50 | 12 May 2023 | 91.6 ± 8.1 | 26.1 ± 9.2 |
| NJ | Sichuan Neijiang | 50 | 5 July 2023 | 104.8 ± 5.7 | 39.7 ± 6.7 |
| DY | Sichuan Daying | 50 | 7 July 2023 | 93.0 ± 4.3 | 25.1 ± 2.7 |
| Locus | Primer Sequence (5′-3′) | Repeat Motif | Ranges (bp) | Tm (°C) | Fluorochromes |
|---|---|---|---|---|---|
| PCL02 | F:ATCAAATCAAACGAAGCAAGAAAG R:GAAGACGGGACACCACGAG | (TG)30 | 245–271 | 47 | TAMRA |
| PC2 | F:ATTGGAAGACCGGGTTCGAGGG R:GGCGTAGAGGAGGTGGTGGT | (CAC)7 | 210–228 | 52 | FAM |
| PC17 | F:TCGTTTCTCCTGTATATCTACCGAGC R: ACACACCAGGCCAGGTCCATCTT | (ATA)9 | 187–211 | 58 | TAMRA |
| PC19 | F:CAGATCAGATTTGCTATGCAGTGTTGTGT R:AGGAATCTAATTGCTTATTCATTGCCTCC | (GCT)12 | 158–191 | 52 | FAM |
| PclG03 | F:AAGCTTACAATAAATATAGATAGAC R:CTCTCCACCAGTCATTTCTT | (TCTA)20 | 216–420 | 52 | HEX |
| PclG07 | F:CCTCCCACCAGGGTTATCTATTCA R: GTGGGTGTGGCGCTCTTGTT | (TCTA)8 | 100–160 | 58 | TAMRA |
| PclG08 | F:ACGATAAATGGATAGATGGATGAA R: CCGGGTCTGTCTGTCTGTCA | (GATA)16 | 148–220 | 58 | HEX |
| PclG09 | F: TATGCACCTTTACCTGAAT R: TGTTGGTGTGGTCATCA | (TCTA)14 | 80–160 | 47 | FAM |
| PclG13 | F:CTCTCCTGGCGCTGTTATTTAGC R:TGAAGAGGCAGAGTGAGGATTCTC | (TCTA)12 | 130–150 | 58 | HEX |
| PclG17 | F: GTC GGG AAC CTA TTT ACA GTG TAT R: AAG AGC GAA GAA AGA GAT AAA GAT | (TCTA)14 | 156–190 | 58 | TAMRA |
| PclG29 | F: GAA AGTCATGGGTGT AGG TGT AAC R: TTT TTG GGC TAT GTG ACG AG | (TATC)9 | 95–165 | 52 | HEX |
| PclG33 | F:TTCGAGGCGTTGCTGATTGTAAGT R: CAAGGAAGCGTATAGCCGGAGTCT | (GT)21 | 120–180 | 58 | FAM |
| Population | Na | Ne | Ho | He | PIC | I | AR | FIS |
|---|---|---|---|---|---|---|---|---|
| QJ * | 5.167 | 2.380 | 0.614 | 0.592 | 0.525 | 0.996 | 4.365 | −0.175 |
| JL * | 4.500 | 2.466 | 0.629 | 0.565 | 0.494 | 1.012 | 4.275 | −0.195 |
| Hubei Mean | 4.834 | 2.423 | 0.622 | 0.578 | 0.509 | 1.004 | 4.320 | - |
| NX | 5.167 | 2.603 | 0.614 | 0.592 | 0.525 | 1.100 | 4.827 | −0.081 |
| QL | 4.580 | 2.481 | 0.636 | 0.575 | 0.509 | 1.047 | 4.379 | −0.187 |
| LX | 4.000 | 2.287 | 0.549 | 0.547 | 0.467 | 0.939 | 3.778 | −0.140 |
| XW | 4.750 | 2.681 | 0.631 | 0.604 | 0.531 | 1.091 | 4.493 | −0.126 |
| NJ | 4.917 | 2.553 | 0.645 | 0.595 | 0.522 | 1.068 | 4.423 | −0.138 |
| DY | 5.250 | 2.646 | 0.699 | 0.607 | 0.535 | 1.121 | 4.875 | −0.261 |
| Sichuan Mean | 4.777 | 2.542 | 0.629 | 0.587 | 0.515 | 1.061 | 4.462 | - |
| QJ * | NX | QL | LX | XW | NJ | DY | JL* | |
|---|---|---|---|---|---|---|---|---|
| QJ * | - | 4.138 | 4.969 | 67.685 | 21.680 | 25.208 | 217.141 | 4.287 |
| NX | 0.057 | - | 2.302 | 5.584 | 4.559 | 5.879 | 3.549 | 18.632 |
| QL | 0.048 | 0.098 | - | 5.332 | 5.167 | 5.674 | 4.449 | 2.045 |
| LX | 0.004 | 0.043 | 0.045 | - | 4.345 | 12.846 | 34.472 | 5.056 |
| XW | 0.011 | 0.052 | 0.046 | 0.010 | - | 18.078 | 16.665 | 4.026 |
| NJ | 0.010 | 0.041 | 0.042 | 0.019 | 0.014 | - | 17.671 | 4.621 |
| DY | 0.001 | 0.066 | 0.053 | 0.007 | 0.015 | 0.014 | - | 4.039 |
| JL* | 0.055 | 0.013 | 0.109 | 0.047 | 0.058 | 0.051 | 0.058 | - |
| Source of Variation | Degree of Freedom | Sum of Squares | Mean of Squares | Variance Components | Percentage of Variation | F-Statistics | p-Value |
|---|---|---|---|---|---|---|---|
| Among groups | 1 | 11.307 | 11.307 | 0.004 Va | 0.137 | FCT = 0.002 | 0.505 (Va and FCT) |
| Among populations within groups | 6 | 76.218 | 12.703 | 0.081 Vb | 3.165 | FSC = 0.042 | <0.0001 (Vb and FSC) |
| Among individuals Within populations | 392 | 804.820 | 2.053 | 0.304 Vc | 11.910 | FIS = 0.163 | <0.0001 (Vc and FIS) |
| within individuals | 400 | 1142.000 | 2.855 | 2.167 Vd | 84.788 | FIT = 0.117 | <0.0001 (Vd and FIT) |
| Total | 799 | 2034.345 | 2.556 | 100 |
| Population | QJ * | JL * | NX | QL | LX | XW | NJ | DY | |
|---|---|---|---|---|---|---|---|---|---|
| H deficiency | TPM | 0.339 | 0.604 | 0.367 | 0.689 | 0.604 | 0.661 | 0.661 | 0.170 |
| H excess | TPM | 0.689 | 0.425 | 0.661 | 0.334 | 0.425 | 0.368 | 0.367 | 0.849 |
| Wilcoxon Test probability H excess or deficiency | TPM | 0.677 | 0.850 | 0.733 | 0.677 | 0.850 | 0.733 | 0.733 | 0.340 |
| Model shift test | L | L | L | L | L | L | L | L | |
| G-W value | 0.549 | 0.566 | 0.596 | 0.592 | 0.602 | 0.581 | 0.527 | 0.643 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, C.; Liao, Q.; Ren, Y.; Shao, W.; Li, Y.; Yang, S.; Du, X.; Wu, J. Genetic Characterization of Procambarus clarkii Cultured in Sichuan Province Based on Microsatellite Markers. Fishes 2024, 9, 419. https://doi.org/10.3390/fishes9100419
Deng C, Liao Q, Ren Y, Shao W, Li Y, Yang S, Du X, Wu J. Genetic Characterization of Procambarus clarkii Cultured in Sichuan Province Based on Microsatellite Markers. Fishes. 2024; 9(10):419. https://doi.org/10.3390/fishes9100419
Chicago/Turabian StyleDeng, Changwen, Qingqing Liao, Yingying Ren, Wuyuntana Shao, Yunkun Li, Shiyong Yang, Xiaogang Du, and Jiayun Wu. 2024. "Genetic Characterization of Procambarus clarkii Cultured in Sichuan Province Based on Microsatellite Markers" Fishes 9, no. 10: 419. https://doi.org/10.3390/fishes9100419
APA StyleDeng, C., Liao, Q., Ren, Y., Shao, W., Li, Y., Yang, S., Du, X., & Wu, J. (2024). Genetic Characterization of Procambarus clarkii Cultured in Sichuan Province Based on Microsatellite Markers. Fishes, 9(10), 419. https://doi.org/10.3390/fishes9100419






