Abstract
This study aims to explore the spatial distribution of false kelpfish (Sebastiscus marmoratus) in the mussel farming area, artificial reef areas of Gouqi Island (Shengsi, China), and natural areas using eDNA detection methods. Surface and bottom water samples were collected at 12 stations in November 2022 and April 2023, totaling 52 samples. We used species-specific primers and probes for quantitative PCR (qPCR) detection of Sebastiscus marmoratus eDNA. The eDNA concentrations differed seasonally (p < 0.05) and did not differ (p > 0.05) among the three sampling areas and two water layers. The greatest eDNA concentrations occurred in the surface layer during the spring. Higher concentrations of Sebastiscus marmoratus eDNA were also found in the mussel aquaculture area. Temperature exhibited a significant positive correlation with Sebastiscus marmoratus eDNA concentration (p < 0.05). Additionally, we developed linear equations predicting the relationship between environmental factors and environmental factors, providing a reference for future fishery resource surveys.
Key Contribution:
This study analyzed the spatial distribution differences of Sebastiscus marmoratus across different seasons and water layers in mussel farming areas, artificial reef areas, and natural waters, as well as the relationship between the environmental DNA concentration of Sebastiscus marmoratus and environmental factors. The research provides crucial insights for developing conservation strategies for Sebastiscus marmoratus and lays the groundwork for further studies.
1. Introduction
The drastic reduction in Earth’s biodiversity has led to irreparable losses in both ecological functionality [1] and the human environment, amounting to an increasingly prominent ecological crisis in the 21st century. Marine biodiversity is a critical component of this global diversity, and due to excessive fishing and environmental pollution in coastal areas, marine ecosystems have seen negative impacts due to human exploitation of the marine environment.
Knowledge about the distribution and abundance of key species is crucial for understanding the functioning of marine ecosystems and implementing effective fisheries resource management [2]. Traditionally, the acquisition of aquatic organism distribution data relies on a series of standardized field survey activities aimed at accurately recording the actual quantity and locations of target species to derive scientific results. However, traditional survey methods have shown several limitations in practice. First, they often require a lot of human and material resources, leading to time-consuming and expensive surveys. Second, due to the differences in sampling environments and the ecological habits of different study subjects, it is sometimes necessary to develop and apply targeted sampling strategies and methods, which can increase the complexity and difficulty of the investigation [3]. Moreover, for fish inhabiting specific habitats, such as reefs, traditional methods are often ineffective and inaccurate for assessing fish distribution due to factors such as habitat complexity and difficulty of approach. In recent years, researchers have used acoustic methods combined with traditional trawling to investigate the biomass of specific fish species before and after the deployment of artificial fish reefs in marine reserves, which has mitigated some disadvantages of traditional survey methods but still has the drawback of complexity [4]. Therefore, it is necessary to explore and develop more advanced and efficient survey methods to meet the needs of aquatic organism distribution monitoring under different environmental conditions; eDNA is a novel ecological monitoring technology where DNA fragments from tissues, such as hair, skin, and scales shed by organisms during their life processes persist in the environment for a period of time. Analysis via high-throughput sequencing or PCR can provide insights into the species identity and quantity of these organisms. Previous studies have shown that the real-time quantitative polymerase chain reaction (qPCR) quantification of eDNA concentration is positively correlated with the biomass of target species, which can include fish [5,6,7,8,9,10]. Hence, eDNA concentration can reflect the abundance or biomass of target organisms. The use of eDNA technology for biological resource monitoring can cover a wide range of areas in a relatively short time period without disturbing the target species or their habitats [11,12]. Additionally, compared to traditional monitoring, eDNA allows for easier sampling. This method is currently being applied to endangered species [13,14], the monitoring of invasive species [15,16], and general biodiversity monitoring [17].
The false kelpfish (Sebastiscus marmoratus) is a reef-dwelling fish species found along the coasts of Japan, Korea, the Philippines, and China’s Bohai, Yellow, and East China Seas, as well as to the east of the Leizhou Peninsula [18]. The false kelpfish (Sebastiscus marmoratus) is a primary catch in recreational fisheries, with peak production during fishing moratoriums (May to August in the East China Sea) reaching up to 55 tons at Shengshan Island and Gouqi Island, accounting for 66% of the annual yield. Ecologically, the false kelpfish (Sebastiscus marmoratus) also plays a crucial role in energy transfer within the marine ecosystem of Gouqi Island. Due to its preference for habitats near reefs, it is highly favored by anglers. This fish species is rarely caught by general trawling (Single Trawler or Double Trawler) and, instead, is captured by fishing, crab pots, and gill nets. Its capture with more labor-intensive sampling methods in part contributes to its high market price.
Existing survey methods and predictions suggest that Sebastiscus marmoratus is widely distributed near reefs, but the specific distribution is not known. Further exploration of the resource distribution of S. marmoratus is beneficial for understanding its ecological habits and implementing scientific fisheries management. There is already an ongoing research basis for monitoring S. marmoratus eDNA, with experiments conducted by Tianxiang et al. [19] showing a strong positive correlation between S. marmoratus aquaculture density and the duration and eDNA concentration of indoor water tanks. This indicates that eDNA concentration can be used to understand and predict the spatial distribution of S. marmoratus to some extent. This study aims to analyze the spatial distribution of S. marmoratus and its seasonal variation using eDNA methods, providing technical support for population surveys, including among populations that are difficult to monitor.
2. Materials and Methods
2.1. Sample Collection
The sampling area (Figure 1) is primarily located around Gouqi Island and is divided into three main zones: the mussel farming area, the artificial reef area, and the natural area. Gouqi Island Marine Reserve has open nearshore waters rich in nutrients as well as abundant reefs and biological resources. The surrounding waters of Gouqi Island are the main area for mussel (Mytilus galloprovincialis) cultivation in Zhejiang Province, China with the aquaculture area nearly doubling from 2003 to 2016. By 2019, the mussel farming area had reached 10 km2, with an output exceeding 180,000 metric tons (SI units), ranking third in the country. The mussel farming area is characterized by the presence of mussel cultivation structures and dense clusters of mussel strings. From 2010 to 2015, a total area of 0.5 km2 of artificial fish reefs (a square reef made of cement) were deployed in the marine reserve [20]. The mussel farming area is characterized by the presence of mussel cultivation structures and dense clusters of mussel strings. Research has shown that the similarity of fish community structure and richness between artificial fish reef areas and natural sea areas is relatively high, suggesting that these artificial reefs may be playing a positive role in fish aggregation and enhancement [21]. So, the artificial reef area features man-made structures designed to enhance marine habitats, often used to attract fish and other marine life. The natural area represents an unaltered, natural marine environment without human-made cultivation or enhancement structures.
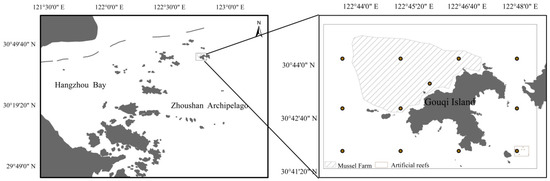
Figure 1.
Sampling station map (the circles on the right represent the locations of the 12 sampling points, the diagonal shadow represents the mussel farming area, the dotted shadow represents the artificial reef area, and the blank background points are the natural areas).
A total of 52 environmental samples were collected in November 2022 and April 2023 in this marine area, from 12 sampling points: natural areas (7), mussel farming areas (4), and artificial reef areas (1). The distance between units is between 2 km and 2.8 km and there is no clustering of similar type sites per regional site. The mussel farming area has its sampling points concentrated in the main regions of the farming zone, with mussel strings densely distributed between the sampling points. In contrast, the natural area, which is a natural sea area, does not have cluster sampling points but, instead, is distributed around the nearshore areas of Gouqi Island. As the artificial reef area is relatively small, sampling was conducted twice at the same location. Sampling times were chosen during low tide each month, and sampling occurred only on sunny days. Surface and bottom seawater samples were collected using a water sampler and SBE 911 Plus CTD system (Sea Bird Scientific, Washington, DC, USA). Surface seawater was obtained at a depth of 1 m and bottom seawater samples were collected at depths between 7 and 30 m. Each seawater sample was collected in 2 L volumes and stored in single-use sterile bags. To measure nutrient content, 1 L of water was collected at each season and each sampling point in the surface water layer. The entire sampling process was completed within 5 h. After sampling, the samples were stored at −20 °C, and filtration was completed within 24 h. Subsequently, 2 L of seawater from each sample was filtered through 47 mm diameter glass fiber filters (0.45 μm pore size) using a vacuum pump in a nearby laboratory. To minimize cross-contamination, filter funnels were rinsed with tap water, immersed in sodium hypochlorite solution for 10 min, rinsed with tap water again, and finally rinsed with distilled water before filtering each sample. Distilled water and tap water were used as negative controls. The entire filtration process was completed within 24 h after sampling. All filtrates, frozen and stored at −196 °C in liquid nitrogen, were transported to the laboratory and stored at −80 °C until further eDNA extraction.
2.2. DNA Extraction
Due to the presence of a large number of coarse particles and sediment in the water sample that were not effectively removed before filtration, they accumulated on the filter membrane, prompting the use of a soil-specific kit for DNA extraction. The eDNA was extracted from filters using the SoilDNAKIT (D5625Omega) kit following the manufacturer’s extraction protocol. The extracted DNA was then stored at −20 °C until required for subsequent PCR analysis.
2.3. QPCR
Quantitative PCR (qPCR) to detect S. marmoratus eDNA was performed using the LightCycler 480 II fluorescence quantitative PCR instrument (Roche, Rotkreuz, Switzerland). The primers and probes used in this experiment were designed based on the COI gene sequence of the false kelpfish (Sebastiscus marmoratus) and the specificity of the primers was verified. The forward primer sequence was H-F1-5′-CTGTTCTAATTACCGCTGTCCTT-3′, the reverse primer sequence was H-R1-5′-CGGCTGGGTCAAAGAATGT-3′, the primer length was 111 bp, and the TaqMan™ probe sequence was HPro-6524: TCTCCCTACCAGTTCTTGCTGCTGGCAT-5′FAM-3′BHQ1, designed to specifically bind to the target region. Sequencing of three positive PCR products was successfully conducted, and the obtained sequences were subjected to BLAST search in the NCBI database. They matched perfectly with the COI sequence of Sebastiscus marmoratus, confirming that the amplified sequences in this experiment are from Sebastiscus marmoratus. The primers, probe, and sequencing were synthesized by Sangon Biotech (Shanghai, China).
Each 20 µL PCR reaction mixture consisted of 2 µL of template DNA extract, 900 nM each of forward and reverse primers, 125 nM of probes, and 1× PCR master mix (FastStart Essential DNA Probes master, Roche Diagnostics, Basel, Switzerland). The amplification conditions were as follows: initial denaturation at 94 °C for 3 min, followed by denaturation at 94 °C for 15 s, annealing at 57 °C for 15 s, and extension at 72 °C for 30 s. After completing these steps, the 96/384-well plate containing the prepared samples was placed in the LightCycler 480 II instrument (Roche, Basel, Switzerland) for amplification. Three replicates of PCR were performed for each water sample (i.e., each DNA extract). Additionally, triplicates of four concentrations of standards were amplified, including artificial DNA fragments with 3 × 101, 3 × 102, 3 × 103, and 3 × 104 copies inserted into the 129 bp target region. The DNA concentration was quantified based on the calibration curve obtained from the standard results. To check for cross-contamination during the PCR process, three non-template controls were included in each 96-well reaction plate.
2.4. Acquisition of Environmental Factor Data
In this study, the environmental factors included temperature, salinity, sampling station depth, chlorophyll, and nutrient concentrations, including NH3−, NO2−, NO3−, and PO43−. These environmental factors were collected in each season, and only surface water layers were collected except for the depth of sampling. Temperature, salinity, station depth, and chlorophyll were obtained using a CTD device during field sampling. The concentrations of NH3−, NO2−, NO3−, and PO43− in seawater were determined using a continuous flow injection method with a continuous flow analyzer (SEAL Analytical AS4 V001, SEAL Analytical Instruments (Shanghai) Co., Ltd., Shanghai, China).
2.5. Data Analysis Methods
To understand the relationship between the eDNA concentration of Sebastiscus marmoratus and the sampling area, water layer, and season, a linear mixed effects model was constructed. In this model, eDNA concentration served as the response variable, while sampling area, water layer, and season were treated as fixed effects, and sampling points were considered random effects. This analysis was performed using the R software version 4.1.3 and the lmer function of the LME4 package [22]. The model residuals did not meet the parametric assumptions and the fish eDNA concentrations were log-transformed (log(x + 1)) before analysis. The formula for the model is as follows:
y = β0 + a × β1 + b × β2 + c × β3 + (1∣site)
Among them,
- y is eDNA concentration;
- Β0 is the intercept;
- the sampling seasonal variable is a;
- the sampling layer variable is b;
- the sampling area variable is c;
- (1|Site) indicates the sampling point as a random effect.
Kriging interpolation was applied to grid the false kelpfish (Sebastiscus marmoratus) eDNA concentration data at each sampling site, and a mask was used to exclude grid data from the study area. It can be used to show qualitative trends in Sebastiscus marmoratus eDNA concentration across different seasons, layers, and areas, and this visualization was completed using ArcGIS 10.8 software.
Correlation analyses between environmental variables were calculated using the “Pheatmap” package [23] and the cor function in R software version 4.1.3 [24].
In the study, multiple linear regression (MLR) models were used to describe the relationships between various environmental factors, allowing for the evaluation of the strength of these relationships and the overall explanatory power of the model. This analysis involves the use of sampling points that were repeatedly sampled, and as such, it is pseudoreplicated. Linear mixed effects model analysis would be ideal; however, we used multiple linear regression analysis to ensure our resulting predictive equations could be used and understood by a greater number of fishery professionals. The R software version 4.1.3 was employed to perform the MLR analysis on data obtained from eDNA concentrations and environmental factors. The specific method is to first construct all models containing 1 to 8 variables, then remove models that include environmental factors with moderate to strong multicollinearity (|r| > 0.5), and finally select the best model based on R2 and RMSE values. The MLR model was constructed using the stats package in R software version 4.1.3 and the lm function.
3. Results
3.1. Plasmid Construction
The amplicon length of the quantified plasmid is 111 bp, and the size of the plasmid is 2803 bp. The concentration of the plasmid standard is 178.5 ng/μL. In the qPCR results, the Ct values of the plasmid standard range from 14.3 to 32. The coefficient of determination (R2) of the curve is 99.99%. The amplification efficiency is 109.7%. Within 40 cycles, only samples with false kelpfish (Sebastiscus marmoratus) DNA as the template were detected, with a detection limit of 31.9, while no other species or negative controls were detected. The results indicate that within the standard range of diluted plasmids, there is a consistent and predictable relationship between the fluorescent signal and the concentration of the target DNA. This indicates stable PCR amplification efficiency, making it effective for quantitative analysis of samples, and the established standard curve accurately reflects the amplification of COI.
3.2. The Concentration of False Kelpfish (Sebastiscus marmoratus) eDNA
The results of the linear mixed effects model show that the cross-validation mean square error is 0.89. The intercept of the model, the estimates of the fixed effects, the standard deviation, and the t-value are shown in Table 1. The eDNA concentrations were significantly affected by sampling season (|t| ≥ 2.58) but less by sampling area and water layer (|t| < 1.96). Figure 2 shows the average Edna concentration in different seasons.

Table 1.
Linear mixed-effects model effect parameter.
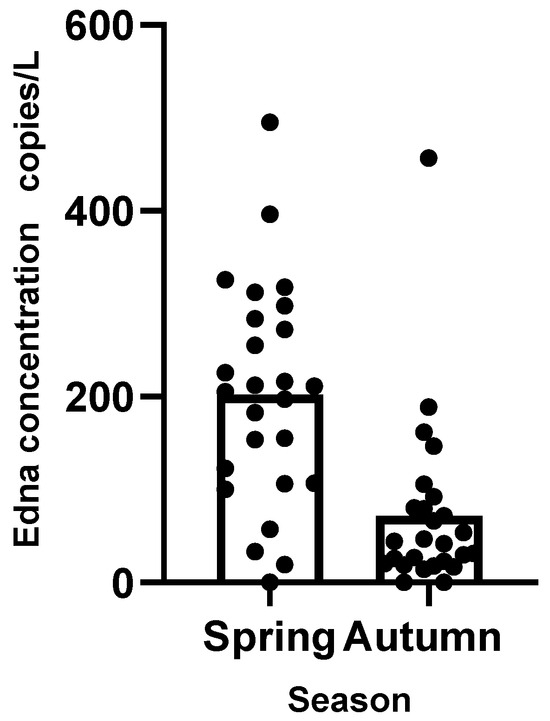
Figure 2.
Mean eDNA concentrations in spring and fall.
Overall, out of 52 eDNA samples, only 3 samples did not detect S. marmoratus DNA, located, respectively, in the northeast outside the mussel aquaculture area in spring and autumn (2 samples) and southwest of Gouqi Island in autumn (1 sample). High eDNA concentrations (copies/L > 300) occurred at five stations in spring and one in autumn, with the highest frequency of occurrence in the mussel aquaculture area (five stations).
The results of the linear mixed effects model show that the cross-validation mean square error is 0.89. The intercept of the model and the estimates of the fixed effects, the standard deviation, and the t-value are shown in Table 1. The eDNA concentrations were significantly affected by sampling season (|t| ≥ 2.58) but less by sampling area and water layer (|t| < 1.96). Figure 2 shows the average eDNA concentration in different seasons. The average eDNA concentration in spring was 202.35 ± 118.75 copies/L, while in autumn it was 71.50 ± 92.54 copies/L.
The visualized heat map shows the spatial and temporal distribution of eDNA concentration in S. marmoratus (Figure 3). The eDNA concentration of S. marmoratus was higher on the east side than on the west side of Gouqi Island in both spring and autumn (Figure 3). In spring, surface eDNA was mainly distributed within the aquaculture area and on both sides of Gouqi Island, with the highest concentration in the center of the aquaculture area (Figure 3); bottom layer eDNA was mainly distributed within the aquaculture area and on the east side of Gouqi Island. In autumn, surface eDNA was mainly distributed in the eastern part of Gouqi Island, with noticeably lower concentrations in the west; bottom layer eDNA was mainly distributed in the northern part of the aquaculture area and the southern part of Gouqi Island, moving toward deeper offshore areas compared to spring.
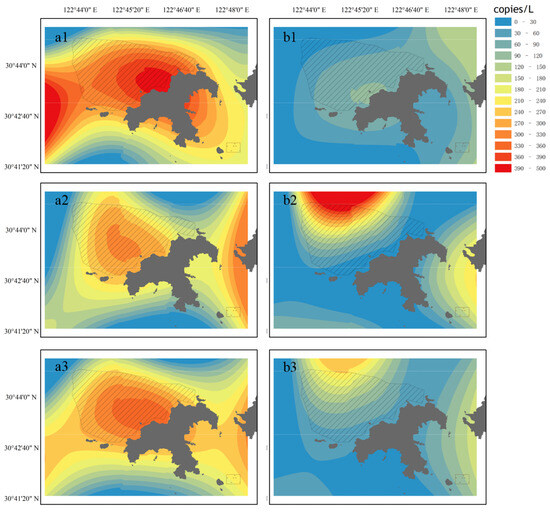
Figure 3.
The eDNA concentration in the sampling area of Sebastiscus marmoratus. (a1) Surface in spring; (a2) bottom in spring; (a3) average in spring; (b1) surface in autumn; (b2) bottom in autumn; (b3) average in autumn.
3.3. The Relationship Between False Kelpfish (S. marmoratus) eDNA Concentration and Environmental Factors
The correlation analysis of eight environmental variables at sampling sites revealed that in spring, the presence and concentration of S. marmoratus eDNA were significantly correlated with temperature, salinity, depth, and nitrite concentration at the sampling points (p < 0.05). However, in autumn, there was no significant correlation between the presence or concentration of S. marmoratus eDNA and any of the environmental factors.
The correlations among independent variables presented in Figure 4 indicate that moderate to strong multicollinearity (|r| > 0.5) occurs between the following: (1) temperature and salinity; (2) NH3− and station depth; (3) salinity and NO2−; (4) salinity and NO3−; (5) NO2− and NO3−; and (6) salinity and PO43−. Models containing combinations of these environmental factors were first excluded, and then the best models were selected based on R2 and RMSE values (Table 2). Additionally, since this study included only 52 samples, only models with fewer than five variables were retained.
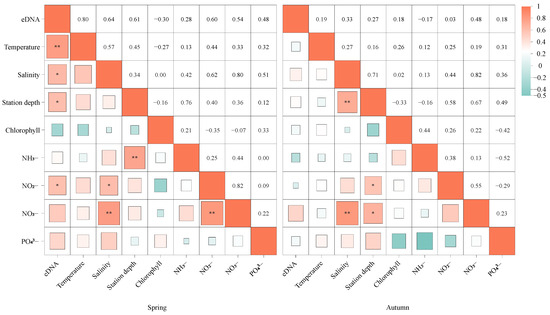
Figure 4.
Correlation between S. marmoratus eDNA concentration and various environmental factors of Sebastiscus marmoratus. The significance level is marked with * (p-value < 0.1), ** (0.05 < p-value < 0.1).

Table 2.
The filtered MLR model.
4. Discussion
4.1. Distribution Characteristics of S. marmoratus
Monitoring eDNA concentrations can help predict the potential distribution of target species across the landscape, reducing workload and research costs and enabling more effective ecological monitoring [10,25]. In this study, eDNA of false kelpfish (S. marmoratus) was detected at all 12 sampling stations, suggesting its widespread distribution in the larger coastal waters near Gouqi Island. Previous research by Zeng Xu et al. [26] on the habitat of false kelpfish (Sebastiscus marmoratus) around Gouqi Island found substantial abundance near Gouqi Island’s coast. Although their survey was limited to within 200 m offshore, their results also demonstrated the species’ dominant status within the local community structure, consistent with findings from this study. The results also confirm the reliability of environmental DNA technology.
The survey results showed that S. marmoratus concentrations were higher in the spring season, which may be closely related to seasonal ecological changes. Spring is typically a season of vigorous reproduction for algae and plankton in marine ecosystems, providing richer food resources and thus leading higher abundance of S. marmoratus in spring. The spawning period of S. marmoratus is from December to April each year, and the survey time of this study was in April, so it is probable that the spawning period of S. marmoratus was coming to an end, and the biomass had reached a high level with fish eggs. Therefore, the high eDNA concentration in spring may be due to the increased biomass resulting from the spawning behavior of S. marmoratus. This conclusion indicates that eDNA surveys can infer fish spawning behavior by monitoring eDNA concentrations. Research by Hayer and Michael et al. [27,28] also confirms this conclusion. Additionally, in autumn, there is a trend for the concentration of S. marmoratus eDNA to move toward deeper offshore areas, similar to the findings of Changwen et al. [29], possibly due to a relative decrease in bait availability near Gouqi Island in autumn, intensifying food competition and leading S. marmoratus to move outward in search of more food sources. (The swimming ability of S. marmoratus juveniles is poor, and they are more susceptible to the influence of ocean currents, so the distribution of eDNA in spring may be larger than the actual range).
In addition to being influenced by their own habits, the distribution of marine fishery resources is also affected by abiotic factors such as physical and chemical factors. Therefore, environmental factors play a key role in fishery resource surveys [30]. According to the MLR model equation obtained, temperature is significantly correlated with S. marmoratus eDNA concentration. Water temperature is one of the main environmental factors affecting fish ecological habits, which not only affects the growth, development, reproduction, and metabolism of fish, but also influences the migratory distribution of fish, the timing of fishing periods, the duration of fishing seasons, the position of central fishing grounds, and the aggregation behavior of fish [31]. Water temperature can also indirectly affect the migratory distribution of fish by influencing the behavior, distribution, and abundance of bait organisms [32]. As the main carrier of phytoplankton photosynthesis, chlorophyll is related to oxygen content in seawater [33]. The content of chlorophyll can reflect the change in plankton abundance, and then reflect the feeding condition of S. marmoratus to some extent; however, plankton abundance may be affected by many other external factors, such as temperature or other low-trophic level fish predation [34,35]. NH3 and NO3 are important sources of nitrogen for aquatic plants. Many Aquatic animals obtain their nitrogen indirectly by feeding on plants or algae. NH3 and NO3 play a key role in the material cycle and energy flow of the aquatic food chain. In ecosystems, the availability of PO4 is often an important factor limiting the growth of aquatic organisms, especially in water bodies with low phosphorus levels, where PO4 supplementation may significantly promote growth. Levels of NH3, NO3, and PO4 should not be too high, as they are also key factors in algal bloom [36].
4.2. Application of eDNA Surveys in Marine Fish: Potential, Limitations, and Prospects
This study reveals the spatial distribution and characteristics of S. marmoratus in the marine reserve of Gouqi Island, including its seasonal variability. The species is widely distributed near Gouqi Island’s coast. Biomass is lower in autumn compared to spring, with a tendency for outward migration. These results can help fishery managers or commercial fisherpeople choose the right fishing time in spring and the right fishing location. In addition, a five-variable combination Edna concentration prediction model was established, which will help fisheries managers predict S. marmoratus abundance. For example, measurements of temperature, chlorophyll, NH3, NO3, and PO4 will help identify high areas of S. marmoratus eDNA and potentially can help catch more S. marmoratus for research purposes or consumption. Also, MLR models suggest that even a simple measurement of temperature and water depth might be sufficient and that false kelpfish appear associated with cooler water temperatures at deeper water depths. Overall, this study helps measure aspects of S. marmoratus behavior and abundance that were difficult to achieve with traditional surveys, such as the distribution of Sebastiscus marmoratus in different water layers. Conventional surveys using trawl nets near the coast are limited by habitats and net height, making it difficult to monitor various water layers and most of the marine area, while also causing disturbances to the ecosystem. Therefore, in some cases, eDNA surveys have higher efficiency compared to traditional methods [37,38,39], such as biodiversity surveys, especially as the cost of genetic research has decreased in recent decades, making genetic-based resource surveys more popular [28]. The advantages of eDNA are evident and will be applied in more research.
However, there are still some issues in current research, particularly the inability to obtain actual survey data for Sebastiscus marmoratus due to the limitations of traditional survey methods. Therefore, it is not possible to accurately assess the correlation between eDNA concentration and the actual biomass of Sebastiscus marmoratus. Additionally, with eDNA technology, we cannot confirm whether the detected species are juveniles or adults, nor can we determine their survival status. Therefore, there is an urgent need for further innovation in eDNA technology to distinguish the sources of eDNA and reveal the basic dynamics of eDNA in the marine environment [40]. Moreover, standardized methods for field surveys and analysis need to be established. Considering the shortcomings in the duration and influencing factors of eDNA signals, eDNA research should be considered as a supplement rather than an independent tool, especially in cases involving marine flow environments [5,41]. Therefore, more research is needed to further improve our understanding of the parameters and processes that may affect eDNA distribution in the ocean.
In recent years, despite the decline in false kelpfish (Sebastiscus marmoratus) resources, certain achievements have been made through restocking and marine reserve construction. Considering the high fecundity of Sebastiscus marmoratus, their production has been maintained to a certain extent. The designation of marine reserves has been proven to play an important role in ecosystem maintenance. To better develop marine reserves, it is necessary to conduct more baseline assessments of ecosystems, such as community structure in mussel farming areas and biomass of various organisms. In these aspects, eDNA technology can play a crucial role.
5. Conclusions
This study utilized eDNA methods to investigate the distribution of S. marmoratus around the marine ranch near Gorgonian Island. The distribution of S. marmoratus exhibited marked seasonal variations. S. marmoratus showed significantly higher abundance in spring compared to autumn due to spawning occurring in the winter and spring. The study of environmental factors revealed a significant correlation between S. marmoratus distribution and temperature. eDNA has proven to be an effective tool for biological monitoring in marine ranches, highlighting the important roles of facilities such as artificial reefs and mussel farms. eDNA holds considerable potential for marine biological monitoring and could be increasingly applied to life history assessments of marine species.
Author Contributions
R.J., H.H. and F.C.: methodology, formal analysis, and writing—original draft preparation. R.Y., P.Z., Y.Z. and X.C.: software and data curation. F.C., Y.Z. and R.J.: writing—review and editing, visualization, supervision, and project administration. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key R & D Program of China (2023YFD2401901).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors express their gratitude to the staff of the Zhejiang Marine Fisheries Research Institute and Key Laboratory of Sustainable Utilization Research for Fishery Resources of Zhejiang Province for help with conducting the experiments.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Valdez, J.W.; Callaghan, C.T.; Junker, J.; Purvis, A.; Hill, S.L.L.; Pereira, H.M. The undetectability of global biodiversity trends using local species richness. Ecography 2023, 2023, e06604. [Google Scholar] [CrossRef]
- Rullens, V.; Stephenson, F.; Lohrer, A.M.; Townsend, M.; Pilditch, C.A. Combined species occurrence and density predictions to improve marine spatial management. Ocean Coast. Manag. 2021, 209, 105697. [Google Scholar] [CrossRef]
- Itakura, H.; Wakiya, R.; Yamamoto, S.; Kaifu, K.; Sato, T.; Minamoto, T. Environmental DNA analysis reveals the spatialdistribution, abundance, and biomass of Japaneseeels at the river-basin scale. Aquat. Conserv. Mar. Freshw. Ecosyst. 2019, 29, 331–520. [Google Scholar] [CrossRef]
- Yuan, X.; Jiang, Y.; Zhang, H.; Jin, Y.; Ling, J. Quantitative assessment of fish assemblages on artificial reefs using acoustic and conventional netting methods, in Xiangshan Bay, Zhejiang Province, China. J. Appl. Ichthyol. 2021, 37, 389–399. [Google Scholar] [CrossRef]
- Yamamoto, S.; Minami, K.; Fukaya, K.; Takahashi, K.; Sawada, H.; Murakami, H.; Tsuji, S.; Hashizume, H.; Kubonaga, S.; Horiuchi, T.; et al. Environmental DNA as a ‘snapshot’ of fish distribution: A case study of Japanese jack mackerel in Maizuru Bay, Sea of Japan. PLoS ONE 2016, 11, e0149786. [Google Scholar]
- Doi, H.; Inui, R.; Akamatsu, Y.; Kanno, K.; Yamanaka, H.; Takahara, T.; Minamoto, T. Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw. Biol. 2016, 62, 30–39. [Google Scholar] [CrossRef]
- Dougherty, M.M.; Larson, E.R.; Renshaw, M.A.; Gantz, C.A.; Egan, S.P.; Erickson, D.M.; Lodge, D.M. Environmental DNA (eDNA) detects the invasive rusty crayfish Orconectes rusticus at low abundances. J. Appl. Ecol. 2016, 53, 722–732. [Google Scholar] [CrossRef]
- Minamoto, T.; Fukuda, M.; Katsuhara, K.R.; Fujiwara, A. Environmental DNA reflects spatial and temporal jellyfish distribution. PLoS ONE 2017, 12, e0173073. [Google Scholar] [CrossRef]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can. J. Fish. Aquat. Sci. 2013, 70, 1123–1130. [Google Scholar] [CrossRef]
- Buxton, A.S.; Groombridge, J.J.; Zakaria, N.B.; Griffiths, R.A. Seasonal variation in environmental DNA in relation to population size and environmental factors. Sci. Rep 2017, 7, 46294. [Google Scholar] [CrossRef]
- Darling, J.A.; Mahon, A.R. From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv Lett 2017, 4, 150–157. [Google Scholar] [CrossRef]
- Feist, S.M.; Jones, R.L.; Copley, J.L.; Pearson, L.S.; Berry, G.A.; Qualls, C.P. Development and validation of an environmental DNA method for detection of alligator snapping turtle (Macrochelys temminckii). Chelonian Conserv. Biol. 2018, 17, 271–279. [Google Scholar] [CrossRef]
- Xu, N.; Xiong, M.H.; Shao, K.; Que, Y.F.; Li, J.Y. Preliminary study on environmental DNA metabarcoding for detecting biodiversity in the middle and lower reaches of the Yangtze River. Res. Environ. Sci. 2020, 33, 1187–1196. [Google Scholar]
- Xu, M.Z.; Yang, Y.; Zhang, J.H.; Fu, X.D. Application Challenges of Environmental DNA Technology in Monitoring the Invasion of Bivalves. Acta Ecol. Sin. 2023, 43, 4423–4433. [Google Scholar]
- Wang, X.Y.; Zhang, H.B.; Lu, G.Q.; Gao, T.X. Detection of an invasive species through an environmental DNA approach: The example of the red drum Sciaenops ocellatus in the East China Sea. Sci. Total Environ. 2022, 815, 152865. [Google Scholar] [CrossRef]
- Gao, T.; Chen, Z.; Wang, X. A New Method for Surveying Fish Diversity in Coastal Areas: Environmental DNA Analysis Technology. J. Zhejiang Ocean Univ. (Nat. Sci. Ed.) 2018, 37, 1–7. [Google Scholar]
- Zhao, S.L.; Xu, H.X.; Zhong, J.S. Fauna of Marine Fishes in Zhejiang; Zhejiang Science and Technology Press: Hangzhou, China, 2016. [Google Scholar]
- Gao, T.-X.; Chen, Z.; Wang, X.-Y.; Zhang, H.-B.; Shi, H.-L. Discussion on the Correlation between Density of Sebastes schlegelii Cultivation and its eDNA. J. Hydrobiol. 2022, 46, 1007–1015. [Google Scholar]
- Pei, W.H.; Hu, C.Y.; Zhang, X.M.; Chen, Y.Y.; Liu, Y.D. Study on the Community Structure of Macrobenthos in the Mussel Breeding Area of Gouqi Island. Oceanol. Et Limnol. Sin. 2023, 54, 526–536. [Google Scholar]
- Li, F.; Xu, M.; Liu, Q.; Wang, Z.F.; Xu, W.J. Ecological restoration zoning for a marine protected area: A case study of Haizhouwan National Marine Park, China. Sci. Total Environ. 2014, 98, 158–166. [Google Scholar] [CrossRef]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf (accessed on 10 April 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Erickson, R.A.; Rees, C.B.; Coulter, A.A.; Merkes, C.M.; McCalla, S.G.; Touzinsky, K.F.; Walleser, L.; Goforth, R.R.; Amberg, J.J. Detecting the Movement and Spawning Activity of Bigheaded Carps with Environmental DNA. Mol. Ecol. Resour. 2016, 16, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhang, S.; Wang, Z.; Lin, J.; Wang, K. Habitat Suitability Assessment for Sebasticus marmoratus in the Saddle Islands. Acta Ecol. Sin. 2016, 36, 3765–3774. [Google Scholar]
- Hayer, C.-A.; Bayless, M.F.; George, A.E.; Thompson, N.; Richter, C.A.; Chapman, D. Use of Environmental DNA to Detect Grass Carp Spawning Events. Fishes 2020, 5, 27. [Google Scholar] [CrossRef]
- Tillotson, M.D.; Kelly, R.P.; Duda, J.J.; Hoy, M.; Kralj, J.; Quinn, T.P. Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol. Conserv. 2018, 220, 1–11. [Google Scholar] [CrossRef]
- Wu, C.W. Biological Research on Sebastiscus marmoratus in the Nearshore Waters of Zhoushan, Zhejiang Province. J. Zhejiang Ocean. Univ. (Nat. Sci. Ed.) 1999, 3, 185–190+226. [Google Scholar]
- Mukherjee, S.; Chaudhuri, A.; Kundu, N.; Mitra, S.; Homechaudhuri, S. Comprehensive analysis of fish assemblages in relation to seasonal environmental variables in an estuarine river of Indian sundarbans. Estuaries Coasts 2012, 36, 192–202. [Google Scholar] [CrossRef]
- Yin, M.L. Fish Ecology; China Agricultural Press: Beijing, China, 1995. [Google Scholar]
- Li, X.D. The Relationship between Seawater Temperature and Fishing Grounds. Acta Oceanol. Sin. 1982, 1, 103–113. (In Chinese) [Google Scholar]
- Belkin, I.M.; O’Reilly, J.E. An algorithm for oceanic front detection in chlorophyll and SST satellite imagery. J. Mar. Syst. 2009, 78, 319–326. [Google Scholar] [CrossRef]
- Andersson, A.; Haecky, P.; Hagström, A. Effect of temperature and light on the growth of micro-nano-and pico-plankton: Impact on algal succession. Mar. Biol. 1994, 120, 511–520. [Google Scholar] [CrossRef]
- Mazumder, A.; Taylor, W.D.; McQueen, D.J.; Lean, D.R. Effects of fish and plankton on lake temperature and mixing depth. Science 1990, 247, 312–315. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.Q. Approaches to Mechanisms and Control of Eutrophication of Shallow Lakes i n the Middle and Lower Reaches of the Yangze River. J. Lake Sci. 2002, 14, 193–202. [Google Scholar]
- Ji, Y.; Ashton, L.; Pedley, S.M.; Edwards, D.P.; Tang, Y.; Nakamura, A.; Kitching, R.; Dolman, P.M.; Woodcock, P.; Edwards, F.A.; et al. Reliable, verifiable and efficient monitoring of biodiversity via meta barcoding. Ecol. Lett. 2013, 16, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.P.; O’donnell, J.L.; Lowell, N.C.; Shelton, A.O.; Samhouri, J.F.; Hennessey, S.M.; Feist, B.E.; Williams, G.D. Genetic signatures of ecological diversity along an urbanization gradient. PeerJ 2016, 4, e2444. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv Genet 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Murakami, H.; Yoon, S.; Kasai, A.; Minamoto, T.; Yamamoto, S.; Sakata, M.K.; Horiuchi, T.; Sawada, H.; Kondoh, M.; Yamashita, Y.; et al. Correction to: Dispersion and degradation of environmental DNA from caged fish in a marine environment. Fish Sci. 2019, 85, 1109. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).