Comparative Transcriptomic Profiling of Brain and Liver in Phoenix Barbs (Spinibarbus denticulatus denticulatus) with Differential Growth Rates
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Animal Samples
2.2. RNA Extraction Sample
2.3. Library Generation and RNA-seq Analysis
2.4. Reverse Transcription Quantitative PCR Analysis
2.5. Statistical Analysis
2.6. Ethical Statement
3. Results
3.1. Growth Performance of FG and SG S. d. denticulatus
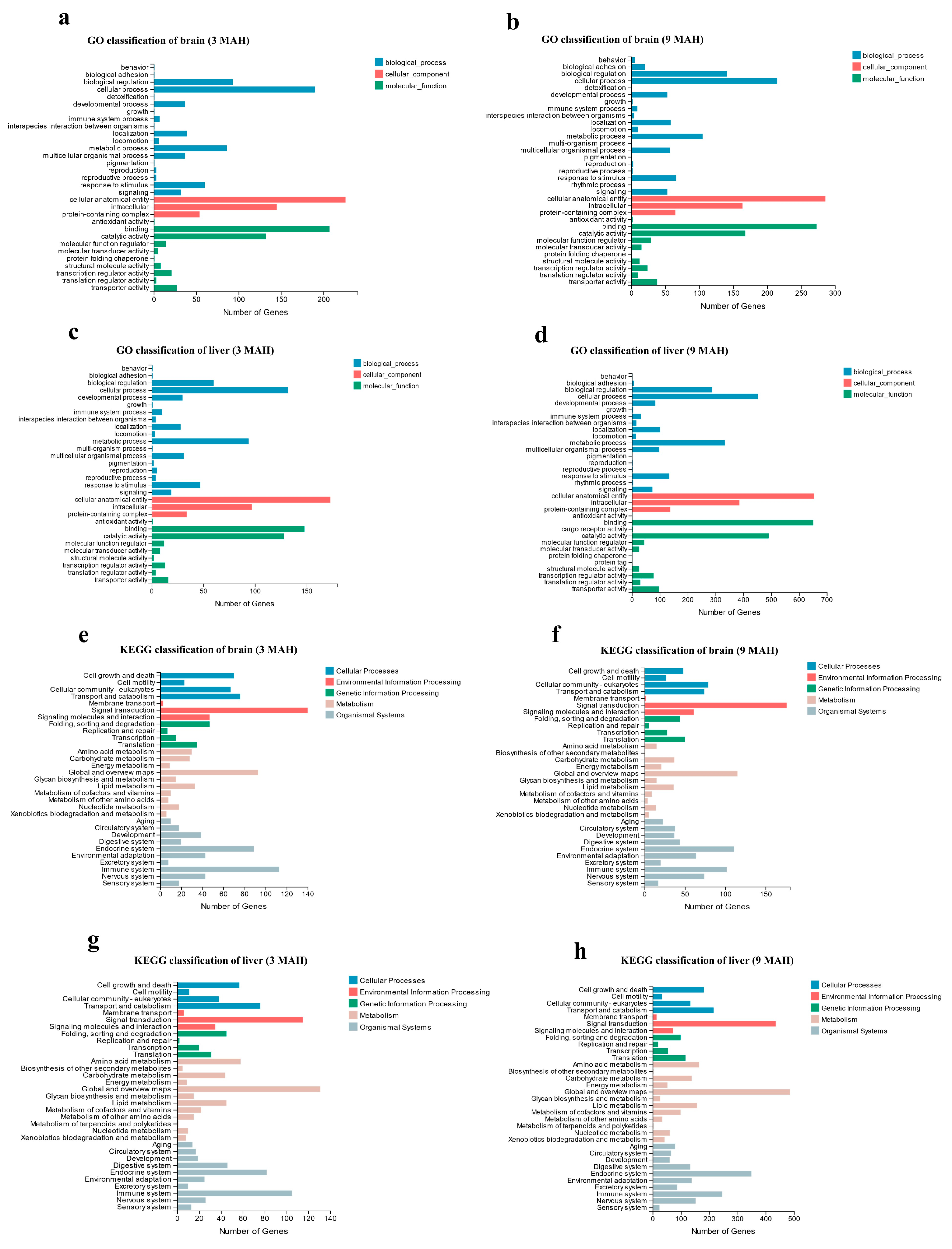
3.2. Sequencing and Annotation
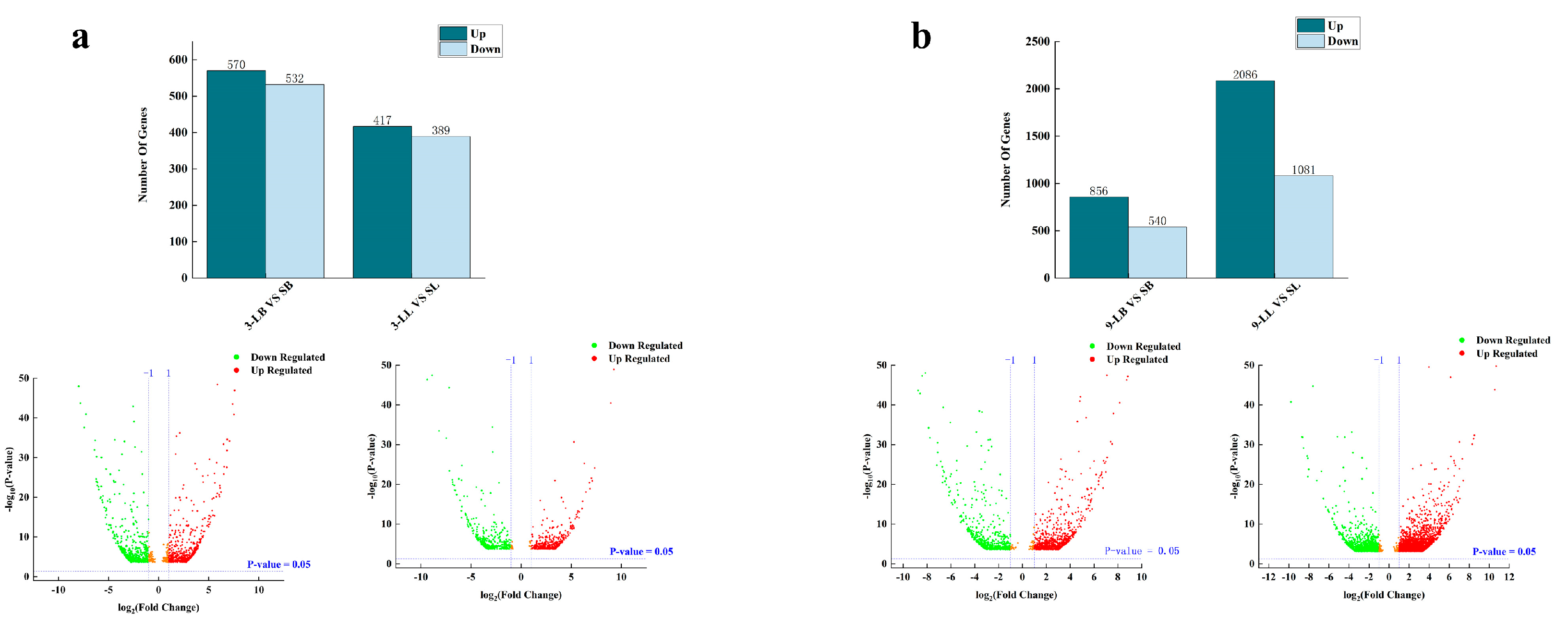
3.3. Identification of DEGs in Liver and Brain Tissues at Two Developmental Stages
3.4. Analysis of DEGs between FG and SG S. d. denticulatus
3.5. Analysis of DEGs between the FG and SG Groups at Two Growth Stages
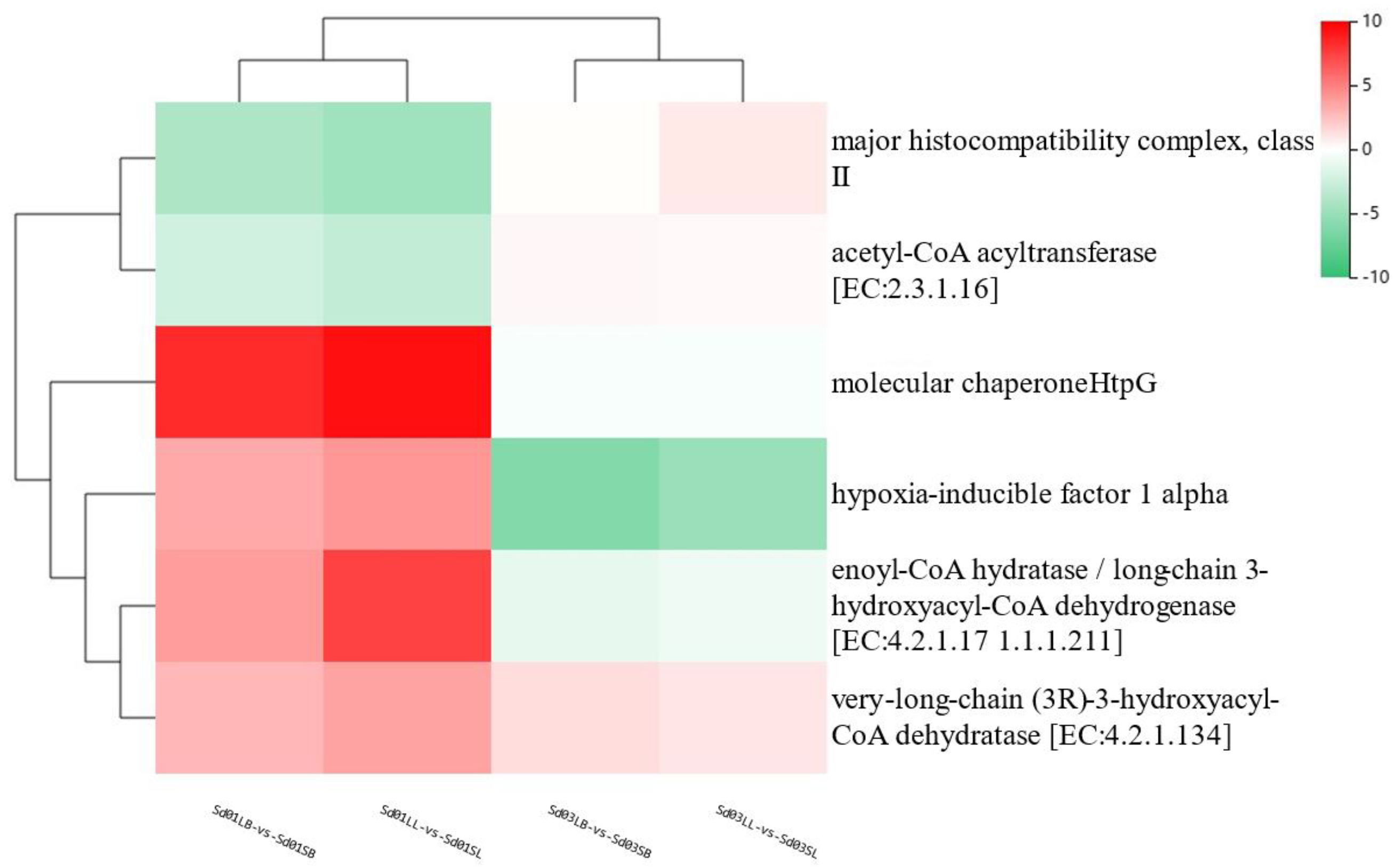
3.6. Analysis of DEGs from the Brain–Liver Axis
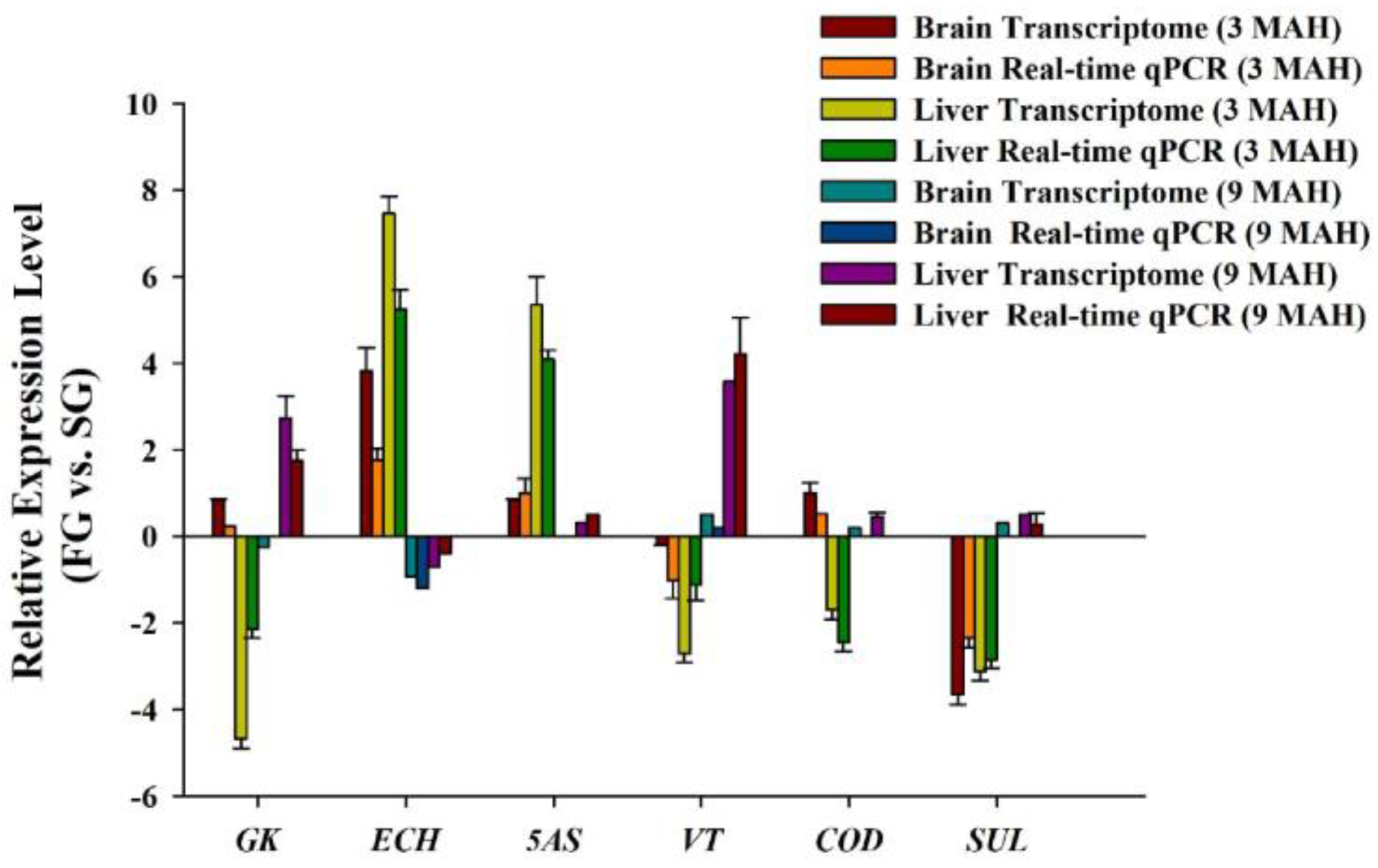
3.7. RT-qPCR Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, X.; Chen, H.M.; Qian, X.Q.; Gui, J.F. Transcriptome analysis of grass carp (Ctenopharyngodon idella) between fast- and slow-growing fish. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100688. [Google Scholar] [CrossRef] [PubMed]
- De-Santis, C.; Jerry, D.R. Candidate growth genes in finfish—Where should we be looking? Aquaculture 2007, 272, 22–38. [Google Scholar] [CrossRef]
- Dai, X.; Zhang, W.; Zhuo, Z.; He, J.; Yin, Z. Neuroendocrine regulation of somatic growth in fishes. Sci. China Life Sci. 2015, 58, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Boyce, W.T.; Sokolowski, M.B.; Robinson, G.E. Genes and environments, development and time. Proc. Natl. Acad. Sci. USA 2020, 117, 23235–23241. [Google Scholar] [CrossRef] [PubMed]
- Gjedrem, T.; Rye, M. Selection response in fish and shellfish: A review. Rev. Aquac. 2018, 10, 168–179. [Google Scholar] [CrossRef]
- Gjedrem, T.; Robinson, N.A. Advances by Selective Breeding for Aquatic Species: A Review. Agric. Sci. 2014, 5, 1152–1158. [Google Scholar] [CrossRef]
- Vieira Ventura, R.; Fonseca, E.S.F.; Manuel Yáñez, J.; Brito, L.F. Opportunities and challenges of phenomics applied to livestock and aquaculture breeding in South America. Anim. Front. 2020, 10, 45–52. [Google Scholar] [CrossRef]
- Houston, R.D.; Bean, T.P.; Macqueen, D.J.; Gundappa, M.K.; Jin, Y.H.; Jenkins, T.L.; Selly, S.L.C.; Martin, S.A.M.; Stevens, J.R.; Santos, E.M.; et al. Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 2020, 21, 389–409. [Google Scholar] [CrossRef]
- Johnston, I.A.; Bower, N.I.; Macqueen, D.J. Growth and the regulation of myotomal muscle mass in teleost fish. J. Exp. Biol. 2011, 214 Pt 10, 1617–1628. [Google Scholar] [CrossRef]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Perelló, M.; Salmerón, C.; Riera-Codina, M.; Ibarz, A.; Fernández-Borràs, J.; Blasco, J.; Capilla, E.; et al. Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture 2017, 467, 28–40. [Google Scholar] [CrossRef]
- Su, J.; Li, W.; Li, H.; Zhou, Z.; Miao, Y.; Yuan, Y.; Li, Y.; Tao, M.; Zhang, C.; Zhou, Y.; et al. Comparative transcriptomic analysis of the brain-liver Axis reveals molecular mechanisms underlying acute cold stress response in Gynogenetic Mrigal carp. Aquaculture 2024, 588, 740908. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, R.; Tian, H.; Qin, X.; Ye, C.; Shi, X.; Xia, C.; Cai, T.; Xie, Y.; Jia, Y.; et al. Sexual dimorphism in Odontobutis sinensis brain-pituitary-gonad axis and liver highlighted by histological and transcriptomic approach. Gene 2022, 819, 146264. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Chi, S.; Wang, J.; Yu, X.; Tong, J. Comparative transcriptomic analyses of brain-liver-muscle in channel catfish (Ictalurus punctatus) with differential growth rate. Comp. Biochem. Physiol. Part D Genom. Proteom. 2024, 49, 101178. [Google Scholar] [CrossRef] [PubMed]
- Degani, G. Somatolactin Transcription during Oogenesis in Female Blue Gourami (Trichogaster trichopterus). Adv. Biol. Chem. 2015, 5, 279–285. [Google Scholar] [CrossRef][Green Version]
- Degani, G. Oogenesis control in multi-spawning blue gourami (Trichogaster trichopterus) as a model for the Anabantidae family. Science 2016, 5, 179–184. [Google Scholar]
- Degani, G.; Schreibman, M. Pheromone of male blue gourami and its effect on vitellogenesis, steroidogenesis and gonadotropin cells in pituitary of the female. J. Fish Biol. 2005, 43, 475–485. [Google Scholar] [CrossRef]
- Degani, G. Effect of sexual behavior on oocyte development and steroid changes in Trichogaster trichopterus (Pallas). Copeia 1993, 1993, 1091–1096. [Google Scholar] [CrossRef]
- Matsubara, Y.; Kiyohara, H.; Teratani, T.; Mikami, Y.; Kanai, T. Organ and brain crosstalk: The liver-brain axis in gastrointestinal, liver, and pancreatic diseases. Neuropharmacology 2022, 205, 108915. [Google Scholar] [CrossRef]
- Fu, B.; Wang, X.; Feng, X.; Yu, X.; Tong, J. Comparative transcriptomic analyses of two bighead carp (Hypophthalmichthys nobilis) groups with different growth rates. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 20, 111–117. [Google Scholar] [CrossRef]
- Fu, B.; Yu, X.; Tong, J.; Pang, M.; Zhou, Y.; Liu, Q.; Tao, W. Comparative transcriptomic analysis of hypothalamus-pituitary-liver axis in bighead carp (Hypophthalmichthys nobilis) with differential growth rate. BMC Genom. 2019, 20, 328. [Google Scholar] [CrossRef]
- Mun, S.H.; You, J.H.; Oh, H.J.; Lee, C.H.; Baek, H.J.; Lee, Y.D.; Kwon, J.Y. Expression Patterns of Growth Related Genes in Juvenile Red Spotted Grouper (Epinephelus akaara) with Different Growth Performance after Size Grading. Dev. Reprod. 2019, 23, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, M.; Schmid, A.; Ermatinger, R.; Loffing-Cueni, D. Insulin-like growth factor I in the teleost Oreochromis mossambicus, the tilapia: Gene sequence, tissue expression, and cellular localization. Endocrinology 1997, 138, 3613–3619. [Google Scholar] [CrossRef] [PubMed]
- Otteson, D.C.; Cirenza, P.F.; Hitchcock, P.F. Persistent neurogenesis in the teleost retina: Evidence for regulation by the growth-hormone/insulin-like growth factor-I axis. Mech. Dev. 2002, 117, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Khalil, W.K.B.; Weiler, U.; Becker, K. Influences of incorporating detoxified Jatropha curcas kernel meal in common carp (Cyprinus carpio L.) diet on the expression of growth hormone- and insulin-like growth factor-1-encoding genes. J. Anim. Physiol. Anim. Nutr. 2013, 97, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Thevasagayam, N.M.; Wan, Z.Y.; Ye, B.Q.; Yue, G.H. Transcriptome Analysis Identified Genes for Growth and Omega-3/-6 Ratio in Saline Tilapia. Front. Genet. 2019, 10, 244. [Google Scholar] [CrossRef]
- Kim, W.K.; Singh, A.K.; Wang, J.; Applegate, T. Functional role of branched chain amino acids in poultry: A review. Poult. Sci. 2022, 101, 101715. [Google Scholar] [CrossRef]
- Vázquez, M.J.; Novelle, M.G.; Rodríguez-Pacheco, F.; Lage, R.; Varela, L.; López, M.; Pinilla, L.; Tena-Sempere, M.; Diéguez, C. AMPK-Dependent Mechanisms but Not Hypothalamic Lipid Signaling Mediates GH-Secretory Responses to GHRH and Ghrelin. Cells 2020, 9, 1940. [Google Scholar] [CrossRef]
- Yin, M.; Zhang, L. Hippo signaling: A hub of growth control, tumor suppression and pluripotency maintenance. J. Genet. Genom. 2011, 38, 471–481. [Google Scholar] [CrossRef]
- Ndandala, C.B.; Zhou, Q.; Li, Z.; Guo, Y.; Li, G.; Chen, H. Identification of Insulin-like Growth Factor (IGF) Family Genes in the Golden Pompano, Trachinotus ovatus: Molecular Cloning, Characterization and Gene Expression. Int. J. Mol. Sci. 2024, 25, 2499. [Google Scholar] [CrossRef]
- Sun, P.; Yin, F.; Tang, B. Effects of Acute Handling Stress on Expression of Growth-Related Genes in Pampus argenteus: Gene expression during stress in pampus. J. World Aquac. Soc. 2017, 48, 166–179. [Google Scholar] [CrossRef]
- Pang, X.; Cao, Z.D.; Fu, S.J. The effects of temperature on metabolic interaction between digestion and locomotion in juveniles of three cyprinid fish (Carassius auratus, Cyprinus carpio and Spinibarbus sinensis). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2011, 159, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Duan, B.; Chen, Q.; Liu, P.; Chi, C.; Yang, H. Studies on Status of Fishery Resources in Three Gorges Reservoir Reaches of the Yangtze River. Acta Hydrobiol. Sin. 2002, 26, 605–611. [Google Scholar]
- Iversen, M.; Finstad, B.; McKinley, R.S.; Eliassen, R.A. The efficacy of metomidate, clove oil, Aqui-S™ and Benzoak® as anaesthetics in Atlantic salmon (Salmo salar L.) smolts, and their potential stress-reducing capacity. Aquaculture 2003, 221, 549–566. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Behera, S.; Swain, S.; Panda, M.K.; Mistri, A.R.; Sahoo, B. Sequence and structural analysis of β- actin protein of fishes, using bioinformatics tools and techniques. Int. J. Biosci. 2014, 4, 249–256. [Google Scholar]
- Liu, Z.J.; Zhu, Z.Y.; Roberg, K.; Faras, A.; Guise, K.; Kapuscinski, A.R.; Hackett, P.B. Isolation and characterization of beta-actin gene of carp (Cyprinus carpio). DNA Seq. 1990, 1, 125–136. [Google Scholar] [CrossRef]
- Dutta, H. Growth in fishes. Gerontology 1994, 40, 97–112. [Google Scholar] [CrossRef]
- Pogoda, H.M.; Hammerschmidt, M. Molecular genetics of pituitary development in zebrafish. Semin. Cell Dev. Biol. 2007, 18, 543–558. [Google Scholar] [CrossRef]
- Sheridan, M.A.; Hagemeister, A.L. Somatostatin and somatostatin receptors in fish growth. Gen. Comp. Endocrinol. 2010, 167, 360–365. [Google Scholar] [CrossRef]
- Nam, B.H.; Moon, J.Y.; Kim, Y.O.; Kong, H.J.; Kim, W.J.; Kim, K.K.; Lee, S.J. Molecular and functional analyses of growth hormone-releasing hormone (GHRH) from olive flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 159, 84–91. [Google Scholar] [CrossRef]
- Bernal, J. Thyroid Hormones in Brain Development and Function. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Carter-Su, C.; Schwartz, J.; Argetsinger, L.S. Growth hormone signaling pathways. Growth Horm. IGF Res. 2016, 28, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Cui, W. Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development 2016, 143, 3050–3060. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B.B. Insulin signaling and action: Glucose, lipids, protein. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Norton, L.; Shannon, C.; Gastaldelli, A.; DeFronzo, R.A. Insulin: The master regulator of glucose metabolism. Metabolism 2022, 129, 155142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, J.; Kopchick, J.J.; Frank, S.J. Disulfide linkage of growth hormone (GH) receptors (GHR) reflects GH-induced GHR dimerization. Association of JAK2 with the GHR is enhanced by receptor dimerization. J. Biol. Chem. 1999, 274, 33072–33084. [Google Scholar] [CrossRef] [PubMed]
- Campbell, G.S. Growth-hormone signal transduction. J. Pediatr. 1997, 131 Pt 2, S42–S44. [Google Scholar] [CrossRef] [PubMed]
- Hakuno, F.; Takahashi, S.I. IGF1 receptor signaling pathways. J. Mol. Endocrinol. 2018, 61, T69–T86. [Google Scholar] [CrossRef] [PubMed]
- Werner, H. The IGF1 Signaling Pathway: From Basic Concepts to Therapeutic Opportunities. Int. J. Mol. Sci. 2023, 24, 14882. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Yu, Y.; Zhang, L.; Dong, L.; Gan, J.; Mao, T.; Liu, T.; Peng, J.; He, L. Comparative analyses of liver transcriptomes reveal the effect of exercise on growth-, glucose metabolism-, and oxygen transport-related genes and signaling pathways in grass carp (Ctenopharyngodon idella). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2021, 262, 111081. [Google Scholar] [CrossRef]
- Boccitto, M.; Kalb, R.G. Regulation of Foxo-dependent transcription by post-translational modifications. Curr. Drug Targets 2011, 12, 1303–1310. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, M.; Sun, Y.; Li, L.; Cheng, P.; Li, X.; Wang, N.; Chen, S.; Xu, W. Identification and Functional Analysis of foxo Genes in Chinese Tongue Sole (Cynoglossus semilaevis). Int. J. Mol. Sci. 2023, 24, 7625. [Google Scholar] [CrossRef]
- Luo, W.; Zhou, Y.; Wang, J.; Yu, X.; Tong, J. Identifying Candidate Genes Involved in the Regulation of Early Growth Using Full-Length Transcriptome and RNA-Seq Analyses of Frontal and Parietal Bones and Vertebral Bones in Bighead Carp (Hypophthalmichthys nobilis). Front. Genet. 2020, 11, 603454. [Google Scholar] [CrossRef] [PubMed]
- Adeva-Andany, M.M.; López-Maside, L.; Donapetry-García, C.; Fernández-Fernández, C.; Sixto-Leal, C. Enzymes involved in branched-chain amino acid metabolism in humans. Amino Acids 2017, 49, 1005–1028. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Functional Roles of Fatty Acids and Their Effects on Human Health. JPEN J. Parenter. Enter. Nutr. 2015, 39 (Suppl. S1), 18s–32s. [Google Scholar] [CrossRef] [PubMed]
- Innis, S.M. Fatty acids and early human development. Early Hum. Dev. 2007, 83, 761–766. [Google Scholar] [CrossRef]
- de Carvalho, C.; Caramujo, M.J. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef]


| 3 MAH | 9 MAH | |||
|---|---|---|---|---|
| FG | SG | FG | SG | |
| Body Weight (g) | 13.88 ± 0.65 c | 11.63 ± 0.36 d | 74.80 ± 2.04 a | 32.23 ± 1.45 b |
| Body Length (cm) | 86.45 ± 1.44 c | 83.40 ± 1.17 d | 166.00 ± 1.63 a | 120.60 ± 3.55 b |
| Body Width (cm) | 13.55 ± 0.30 c | 12.18 ± 1.06 c | 23.95 ± 0.29 a | 17.40 ± 0.43 b |
| Body Height (cm) | 25.60 ± 0.56 c | 23.10 ± 1.73 c | 46.85 ± 0.63 a | 32.10 ± 0.99 b |
| Head Length (cm) | 20.70 ± 0.33 c | 19.80 ± 0.63 c | 33.95 ± 0.28 a | 27.30 ± 0.87 b |
| Tissue | Period | Gene ID | Gene Description | Fold Change | p Value |
|---|---|---|---|---|---|
| Brain | 3 MAH | Th17 cell differentiation | |||
| isoform_86026 | molecular chaperone HtpG | 8.34 | <0.01 | ||
| isoform_378722 | proto-oncogene protein c-fos | 3.26 | <0.01 | ||
| isoform_32558 | splicing factor, arginine/serine-rich 1 | 2.14 | <0.01 | ||
| isoform_132081 | solute carrier family 30 (zinc transporter), member 10 | 1.35 | <0.01 | ||
| IL-17 signaling pathway | |||||
| isoform_354536 | hypoxia-inducible factor 1 alpha | 3.35 | <0.01 | ||
| Estrogen signaling pathway | |||||
| isoform_222321 | GTPase KRas/Thermogenesis | 2.85 | <0.01 | ||
| isoform_242870 | calmodulin | 2.78 | <0.01 | ||
| isoform_268095 | heat shock 70 kDa protein 1/2/6/8 | 2.29 | <0.01 | ||
| NOD-like receptor signaling pathway | |||||
| isoform_270527 | mitochondrial FAD-linked sulfhydryl oxidase | 2.79 | <0.01 | ||
| isoform_132081 | solute carrier family 30 (zinc transporter), member 10 | 1.35 | <0.01 | ||
| isoform_221836 | ryanodine receptor 3 | 1.19 | <0.01 | ||
| Valine, leucine, and isoleucine degradation | |||||
| isoform_296751 | methylmalonyl-CoA mutase | 6.33 | <0.01 | ||
| isoform_38010 | acetyl-CoA acyltransferase 2 | 2.82 | <0.01 | ||
| isoform_228961 | dihydrolipoamide dehydrogenase | 2.67 | <0.01 | ||
| Fatty acid elongation | |||||
| isoform_331832 | long-chain 3-hydroxyacyl-CoA dehydrogenase | 3.84 | <0.01 | ||
| isoform_38010 | acetyl-CoA acyltransferase 2 | 2.82 | <0.01 | ||
| isoform_213152 | very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase | 2.80 | <0.01 | ||
| Propanoate metabolism | |||||
| isoform_296751 | methylmalonyl-CoA mutase | 6.33 | <0.01 | ||
| isoform_331832 | enoyl-CoA hydratase | 3.84 | <0.01 | ||
| isoform_360405 | L-lactate dehydrogenase | 3.25 | <0.01 | ||
| isoform_228961 | dihydrolipoamide dehydrogenase | 2.67 | <0.01 | ||
| PI3K-Akt signaling pathway | |||||
| isoform_73678 | 14-3-3 protein gamma/eta | 2.92 | <0.01 | ||
| isoform_222321 | GTPase KRas | 2.85 | <0.01 | ||
| isoform_232012 | thrombospondin 1 | 2.52 | <0.01 | ||
| isoform_342907 | cytochrome c oxidase assembly protein subunit 11 | 2.16 | <0.01 | ||
| isoform_263042 | GTPase HRas | 2.12 | <0.01 | ||
| Brain | 9 MAH | O-glycan biosynthesis | |||
| isoform_368717 | beta-galactoside alpha-2,6-sialyltransferase | 4.53 | <0.01 | ||
| isoform_168720 | protein O-GlcNAc transferase | 2.73 | <0.01 | ||
| Insulin secretion | |||||
| isoform_32522 | guanine nucleotide-binding protein subunit alpha-11 | 4.92 | <0.01 | ||
| isoform_166285 | pituitary adenylate cyclase-activating polypeptide type I receptor | 3.12 | <0.01 | ||
| isoform_179654 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 2/4 | 3.03 | <0.01 | ||
| isoform_361861 | guanine nucleotide-binding protein subunit alpha-11 | 2.63 | <0.01 | ||
| Thyroid hormone signaling pathway | |||||
| isoform_113388 | hypoxia-inducible factor 1 alpha | 6.64 | <0.01 | ||
| isoform_225372 | mediator of RNA polymerase II transcription subunit 13 | 4.33 | <0.01 | ||
| GnRH signaling pathway | |||||
| isoform_326196 | guanine nucleotide-binding protein | 4.12 | <0.01 | ||
| Sphingolipid signaling pathway | |||||
| isoform_209109 | ceramide synthetase | 3.75 | <0.01 | ||
| Liver | 3 MAH | Glycine, serine, and threonine metabolism | |||
| isoform_48613 | alanine-glyoxylate transaminase | 5.36 | <0.01 | ||
| isoform_321441 | 5-aminolevulinate synthase | 5.23 | <0.01 | ||
| isoform_365726 | L-pipecolate oxidase | 4.51 | <0.01 | ||
| isoform_41453 | dihydrolipoamide dehydrogenase | 3.62 | <0.01 | ||
| isoform_257499 | glycine amidinotransferase | 3.56 | <0.01 | ||
| Propanoate metabolism | |||||
| isoform_296751 | methylmalonyl-CoA mutase | 7.18 | <0.01 | ||
| isoform_41453 | dihydrolipoamide dehydrogenase | 3.62 | <0.01 | ||
| Fat digestion and absorption | |||||
| isoform_220933 | aspartate aminotransferase | 8.18 | <0.01 | ||
| isoform_163445 | fatty acid-binding protein | 3.79 | <0.01 | ||
| isoform_51344 | apolipoprotein | 3.76 | <0.01 | ||
| isoform_317355 | microsomal triglyceride transfer protein | 3.64 | <0.01 | ||
| isoform_293387 | apolipoprotein | 2.02 | <0.01 | ||
| Glycerophospholipid metabolism | |||||
| isoform_170346 | glycerol-3-phosphate dehydrogenase (NAD+) | 4.35 | <0.01 | ||
| isoform_14990 | ethanolamine-phosphate cytidylyltransferase | 3.43 | <0.01 | ||
| isoform_245223 | membrane dipeptidase | 2.42 | <0.01 | ||
| isoform_25396 | glyceronephosphate O-acyltransferase | 2.20 | <0.01 | ||
| isoform_5689 | phosphatidate phosphatase LPIN | 2.08 | <0.01 | ||
| Th17 cell differentiation | |||||
| isoform_86026 | molecular chaperone HtpG | 9.36 | <0.01 | ||
| isoform_125178 | major histocompatibility complex, class II | 5.40 | <0.01 | ||
| isoform_354536 | hypoxia-inducible factor 1 alpha | 4.08 | <0.01 | ||
| IL-17 signaling pathway | |||||
| isoform_8916 | transcription factor | 3.58 | <0.01 | ||
| isoform_32558 | splicing factor, arginine/serine-rich 1 | 2.49 | <0.01 | ||
| Fatty acid elongation | |||||
| isoform_331832 | long-chain 3-hydroxyacyl-CoA dehydrogenase | 7.46 | <0.01 | ||
| isoform_213152 | very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase | 3.63 | <0.01 | ||
| Liver | 9 MAH | Glycerolipid metabolism | |||
| isoform_129084 | triose/dihydroxyacetone kinase | 3.41 | <0.01 | ||
| isoform_173628 | glycerol kinase | 3.36 | <0.01 | ||
| isoform_294807 | glycerol-3-phosphate O-acyltransferase 1/2 | 2.84 | <0.01 | ||
| FoxO signaling pathway | |||||
| isoform_27401 | homer | 3.78 | <0.01 | ||
| isoform_361779 | catalase | 3.18 | <0.01 | ||
| isoform_379101 | 5′-AMP-activated protein kinase | 2.95 | <0.01 | ||
| isoform_327226 | B-Raf proto-oncogene serine/threonine-protein kinase | 2.59 | <0.01 | ||
| isoform_344831 | forkhead box protein O3 | 2.28 | <0.01 | ||
| PPAR signaling pathway | |||||
| isoform_18527 | carnitine O-palmitoyltransferase | 8.63 | <0.01 | ||
| isoform_220131 | long-chain acyl-CoA synthetase | 3.45 | <0.01 | ||
| isoform_173628 | glycerol kinase | 3.36 | <0.01 | ||
| isoform_33276 | cholesterol 7 alpha-monooxygenase | 3.26 | <0.01 | ||
| isoform_20614 | spermine oxidase | 2.48 | <0.01 | ||
| Glycine, serine, and threonine metabolism | |||||
| isoform_37652 | sarcosine oxidase/L-pipecolate oxidase | 8.53 | <0.01 | ||
| isoform_170963 | guanidinoacetate N-methyltransferase | 4.37 | <0.01 | ||
| isoform_66322 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | 3.63 | <0.01 | ||
| isoform_323882 | betaine-homocysteine S-methyltransferase | 2.83 | <0.01 | ||
| AMPK signaling pathway | |||||
| isoform_66227 | elongation factor 2 | 11.54 | <0.01 | ||
| isoform_18527 | carnitine O-palmitoyltransferase 1 | 8.63 | <0.01 | ||
| isoform_117486 | ribosomal protein S6 kinase beta | 3.64 | <0.01 | ||
| isoform_309342 | hydroxymethylglutaryl-CoA reductase | 3.44 | <0.01 | ||
| isoform_379101 | 5′-AMP-activated protein kinase | 2.95 | <0.01 | ||
| isoform_344831 | forkhead box protein O3 | 2.28 | <0.01 | ||
| Insulin signaling pathway | |||||
| isoform_12353 | proto-oncogene C-crk | 4.12 | <0.01 | ||
| isoform_117486 | ribosomal protein S6 kinase beta | 3.64 | <0.01 | ||
| isoform_240467 | pyruvate kinas | 3.28 | <0.01 | ||
| isoform_313475 | protein phosphatase 1 regulatory | 3.12 | <0.01 | ||
| isoform_379101 | 5′-AMP-activated protein kinase, regulatory gamma subunit | 2.95 | <0.01 | ||
| isoform_231208 | glucokinase | 2.72 | <0.01 | ||
| isoform_327226 | B-Raf proto-oncogene serine/threonine-protein kinase | 2.59 | <0.01 | ||
| isoform_3120 | phosphorylase kinase | 2.24 | <0.01 | ||
| Period | Gene ID | Gene Description | Fold Change (Brain) | Fold Change (Liver) | p Value |
|---|---|---|---|---|---|
| 3 MAH | Fatty acid elongation | ||||
| isoform_ 119505 | long-chain 3-hydroxyacyl-CoA dehydrogenase | −3.96 | −5.23 | <0.01 | |
| isoform_ 213152 | very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase | 2.80 | 3.63 | <0.01 | |
| isoform_ 331832 | enoyl-CoA hydratase/long-chain 3-hydroxyacyl-CoA dehydrogenase | 3.84 | 7.46 | <0.01 | |
| isoform_ 62084 | acetyl-CoA acyltransferase | −5.76 | −7.35 | <0.01 | |
| Th17 cell differentiation | |||||
| isoform_ 62373 | molecular chaperone HtpG | −9.47 | −9.10 | <0.01 | |
| isoform_ 148791 | signal transducer and activator of transcription 3 | −2.43 | −1.61 | <0.01 | |
| isoform_ 42223 | proto-oncogene protein c-fos | −2.15 | −1.34 | <0.01 | |
| isoform_ 13757 | interleukin 7 receptor | −1.86 | −0.93 | <0.01 | |
| isoform_ 286564 | immunoglobulin lambda-like polypeptide 1 | −1.43 | −3.33 | <0.01 | |
| isoform_ 110868 | non-receptor tyrosine-protein kinase TYK2 | −1.14 | −0.38 | <0.01 | |
| isoform_ 132081 | solute carrier family 30 (zinc transporter), member 10 | 1.35 | 0.00 | <0.01 | |
| isoform_ 354536 | hypoxia-inducible factor 3 alpha | 3.35 | 4.08 | <0.01 | |
| IL-17 signaling pathway | |||||
| isoform_ 39669 | major histocompatibility complex, class II | −3.44 | −4.45 | <0.01 | |
| isoform_ 115641 | molecular chaperone HtpG | −2.79 | −2.31 | <0.01 | |
| isoform_ 135192 | prostaglandin-endoperoxide synthase 2 | −2.49 | −1.56 | <0.01 | |
| isoform_ 125178 | major histocompatibility complex, class II | 1.09 | 5.40 | <0.01 | |
| isoform_ 32558 | splicing factor, arginine/serine-rich 1 | 2.14 | 2.49 | <0.01 | |
| isoform_ 354536 | hypoxia-inducible factor 2 alpha | 3.35 | 4.08 | <0.01 | |
| 9 MAH | Insulin signaling pathway | ||||
| isoform_ 158560 | mitogen-activated protein kinase kinase 2 | −5.76 | −5.63 | <0.01 | |
| isoform_ 189742 | mitogen-activated protein kinase kinase 2 | −4.92 | −5.64 | <0.01 | |
| isoform_ 189742 | mitogen-activated protein kinase kinase 2 | −4.92 | −5.64 | <0.01 | |
| isoform_ 118529 | atrophin-1 interacting protein 1 | −3.84 | −0.06 | <0.01 | |
| isoform_ 30052 | mitogen-activated protein kinase kinase 1 | −3.33 | −1.72 | <0.01 | |
| isoform_ 27740 | exocyst complex component 7 | −1.74 | −2.63 | <0.01 | |
| isoform_ 187116 | hexokinase | −1.38 | −0.43 | <0.01 | |
| isoform_ 107030 | phosphoenolpyruvate carboxykinase | −1.30 | −2.69 | <0.01 | |
| isoform_ 32627 | MAP kinase interacting serine/threonine kinase | −1.19 | −2.67 | <0.01 | |
| isoform_ 357827 | RAF proto-oncogene serine/threonine-protein kinase | −1.17 | −2.60 | <0.01 | |
| isoform_ 125524 | SHC-transforming protein 2 | −1.01 | −2.76 | <0.01 | |
| isoform_ 66303 | protein phosphatase 1 regulatory subunit 3A/B/C/D/E | −1.00 | −3.68 | <0.01 | |
| isoform_ 117486 | ribosomal protein S6 kinase beta | 2.87 | 3.64 | <0.01 | |
| isoform_ 20826 | Rap guanine nucleotide exchange factor 1 | 3.43 | 2.61 | <0.01 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, X.; Zhuang, J.; Liao, X.; Xu, Z.; Liang, W.; Su, Y.; Lin, L.; Xie, J.; Lin, W. Comparative Transcriptomic Profiling of Brain and Liver in Phoenix Barbs (Spinibarbus denticulatus denticulatus) with Differential Growth Rates. Fishes 2024, 9, 411. https://doi.org/10.3390/fishes9100411
Xie X, Zhuang J, Liao X, Xu Z, Liang W, Su Y, Lin L, Xie J, Lin W. Comparative Transcriptomic Profiling of Brain and Liver in Phoenix Barbs (Spinibarbus denticulatus denticulatus) with Differential Growth Rates. Fishes. 2024; 9(10):411. https://doi.org/10.3390/fishes9100411
Chicago/Turabian StyleXie, Xi, Jiamiao Zhuang, Xianping Liao, Zhengsheng Xu, Wenlang Liang, Yilin Su, Li Lin, Jungang Xie, and Weiqiang Lin. 2024. "Comparative Transcriptomic Profiling of Brain and Liver in Phoenix Barbs (Spinibarbus denticulatus denticulatus) with Differential Growth Rates" Fishes 9, no. 10: 411. https://doi.org/10.3390/fishes9100411
APA StyleXie, X., Zhuang, J., Liao, X., Xu, Z., Liang, W., Su, Y., Lin, L., Xie, J., & Lin, W. (2024). Comparative Transcriptomic Profiling of Brain and Liver in Phoenix Barbs (Spinibarbus denticulatus denticulatus) with Differential Growth Rates. Fishes, 9(10), 411. https://doi.org/10.3390/fishes9100411








