Abstract
Xeroderma pigmentosum complementation group C (XPC) protein recognizes bulky DNA adducts to initiate global genomic nucleotide excision repair (GG-NER). Humans carrying germline mutations in the XPC gene display strong susceptibility to skin and certain internal cancers. In addition to its role in NER, recent studies have indicated that XPC is also involved in other DNA damage repair pathways and transcription regulation. In this report, we generated a zebrafish xpc knockout mutant. Zebrafish xpc−/− mutant fish develop relative normally and are fertile. However, the mutant embryos were more sensitive to ultraviolet (UV) irradiation. Upon UV irradiation, compared with the wild type embryos, mutant embryos accumulated significantly higher levels of unrepaired DNA damages and apoptotic cells, which led to more severe abnormal development. Transcriptome analysis showed that the p53 signal pathway and apoptosis were enriched in the over upregulated genes in UV-irradiated mutant embryos, suggesting that high levels of unrepaired DNA lesions activated p53 to trigger apoptotic activity in mutant embryos. More interestingly, up to 972 genes in the untreated mutant embryos were differentially expressed, compared with those in the untreated WT. Among these differentially expressed genes (DEGs), 379 genes did not respond to UV irradiation, indicating that Xpc plays a role in addition of DNA damage repair. Our results demonstrate that Xpc is an evolutionally conserved factor in NER repair. Zebrafish xpc−/− mutant also provides a platform to study other functions of Xpc beyond the DNA damage repair.
Key Contribution:
Our work demonstrate that zebrafish Xpc has evolutionally conserved function in NER repair, and show that Xpc has functions beyond DNA damage response. Zebrafish xpc−/− mutant will provide a plat form to further investigate whether the function of Xpc beyond DNA repair is related to different internal cancers.
1. Introduction
Throughout life, cells are under a wide range of DNA damage pressures from both endogenous and exogenous sources. To minimize the effects of toxic insults on their DNA, cells have evolved different DNA repair pathways. A defect in any DNA repair pathway will result in mutations and genomic instability, which predispose an organism to cancer. The nucleotide excision repair (NER) is the primary DNA repair pathway to eliminate cyclobutane pyrimidine dimers (CPDs) and pyrimidine-pyrimidone (6-4) photoproducts (6-4PPs) generated by ultraviolet (UV) irradiation or DNA strand crosslinking induced by DNA alkylating agents like cisplatin [1,2]. NER has two subpathways: global genomic NER (GG-NER) and transcription coupled NER (TC-NER). Both NER subpathways repair helix-destabilizing DNA lesions. TC-NER occurs when the RNA polymerase II is stalled at bulky DNA lesions within actively transcribed regions of the genome, and GG-NER removes bulky DNA lesions throughout both transcribed and non-transcribed regions of the genome [3,4,5]. In the process of NER, there are four essential steps: recognition, incision/excision, resynthesis and ligation [6]. Two subpathways are different at the step of DNA damage recognition. TC-NER is rapidly triggered by the stagnation of the RNA polymerase II at the site of a DNA damaging lesion, which subsequently recruits CSB and other TC-NER recognition proteins such as CSA, XAB2, UVSSA, USP7 and others [7,8]. GG-NER is initiated by xeroderma pigmentosum group C (XPC), together with RAD23A/B and CETN2, through identification of DNA strand distortions from large, bulky DNA lesions, and is primarily responsible for the slower repair of damage on non-transcribed portions [9,10,11,12]. The following steps after damage recognition are the same between the two NER subpathways. XPC plays an essential role in damage recognition and initiation of GG-NER but is dispensable for TC-NER [13,14].
Xeroderma Pigmentosum (XP) is clinically characterized by photosensitivity and a high incidence of solar irradiation-induced skin tumors and divided into eight different groups (XP-A to XP-G and XP-V). XP-A to XP-G are caused by genetic mutations in one out of seven NER genes, and XP-V derives from mutations in the DNA polymerase-η gene [15]. The extremely high sensitivity of XP patients to sunburn shows that there is a causal relationship between UV-induced erythema and edema with the persistence of UV photoproducts in the DNA, because of the defect of NER [16,17]. XP-C is due to loss-of-function of the XPC protein encoded by the XPC gene. The XPC protein is composed of 940 amino acids and forms a complex with RAD23B and CETN2 to recognize helix distortions on the basis of disrupted base pairing [13]. Xpc−/− knockout mice have also shown an increased susceptibility to UV-induced skin cancer, similar to humans [18].
Several lines of clinical data have shown that loss-of-function of XPC is correlated not only with development of skin cancer but also with development of internal cancers such as lung and bladder [19,20]. Similar observations have been obtained from mouse models. Xpc−/− mice were highly susceptible to lung and liver cancers upon exposure to a single intraperitoneal injection of 2-acetylaminofluorene (2-AAF) and NaOH-2-AAF [21,22]. Interestingly, XPC presents multiple interactions, either physical or regulatory, with the different DNA repair pathways such as base excision repair (BER), mismatch repair (MMR), homologous recombination (HR), and non-homologous end joining (NHEJ) [13,23]. In addition to DNA damage repair, XPC has been reported to be involved in transcription regulation [24,25,26].
In this study, we generated a zebrafish xpc genetic mutant to investigate the Xpc function. Our results showed that the mutant embryos were more sensitive to UV irradiation. Transcriptomic analyses indicated that zebrafish Xpc may also play a role in addition to DNA damage repair. Zebrafish xpc−/− mutant provides a good model to study Xpc functions beyond DNA repair.
2. Materials and Methods
2.1. Phylogenetic Analysis
Six XPC protein sequences including zebrafish and its orthologues in Homo sapiens, Mus musculus, Saccharomyces cerevisiae, Rattus norvegicus and Drosophila melanogaster were collected from NCBI databases [27]. The protein sequences were aligned with MUSCLE. Based on the maximum likelihood algorithm using 1000 replicates of bootstrapping with MEGA version 11, the phylogenetic tree was constructed [28]. The iTOL version 6.5.8 was used for the final editing [29].
2.2. Zebrafish Husbandry and Generation of xpc−/− Zebrafish Mutant
Zebrafish (Danio rerio) AB strain was used as the wild type (WT) in this study. Fish were raised and maintained according to standard procedures described in ZFIN (http://www.zfin.org, accessed on 2 February 2018.).
The xpc−/− knockout zebrafish line was generated by CRISPR/Cas9 according to the published protocol with slight modifications [30]. Briefly, the guide RNA (gRNA: 5′-AGTTGACTCCGAGTGGTGGGAG-3′) was designed to target the eighth exon of the xpc gene by CHOPCHOP [31]. Synthesized gRNA (300 ng/μL) [32] was co-injected with the Cas9 protein (0.25μg/μL) into zebrafish wild type embryos at one-cell stage.
The injected embryos (F0) were raised to adulthood and crossed with WT fish to generate F1. The mutant fish were identified by a 451 bp DNA fragment (containing the gRNA-target site), amplified with a pair of primers (forward 5′-GATGTGGATCAGGGTGTCGG-3′; reverse: 5′-CAAACACTGGAGACACGGCT-3′), followed by BslI endonuclease digestion. The mutations were verified by sequencing.
2.3. Whole-Mount In Situ Hybridization
The xpc RNA probes were amplified with a pair of primers (forward: 5′-TAGATGTGGATCAGGGTGTCGG-3′; reverse: 5′-TCCTGGCTTTACGGGAACGGTT-3′) from the cDNA of zebrafish WT embryos at 24 h post fertilization (hpf). Whole-mount in situ hybridization (WISH) was performed according to the previously described protocol [33].
2.4. Exposure of UV
For each replicate, about 50 WT or xpc−/− mutant embryos at 28 hpf were exposed to UV (254 nm) irradiation in CL-1000 UV crosslinker (BD Biosciences) at the dosage of 75 J/m2 or 180 J/m2.
2.5. Comet Assay
For the comet assay, approximately 50 irradiated or unirradiated control embryos were sampled at 0 and 24 h post irradiation (hpi), and subjected to cell dissociation in ice-cold PBS containing 20 mM EDTA [34]. The comet assay was performed according to the published paper [35] with an alkaline treatment at pH 11. For the data processing, each comet picture was measured with CASP (Comet assay software project).
2.6. TUNEL Assay
UV-irradiated or unirradiated control embryos were sampled at 24 hpi, fixed with 4% PFA overnight and then subjected to the TUNEL assay using the In Situ Cell Death Detection Kit, TMR red (Roche) [36]. After three rinses in PBS, the TUNEL signals were captured by inverted fluorescence microscope (Olympus IX73).
2.7. Transcriptome Analysis
The total RNAs were extracted from pools of 30 zebrafish embryos of WT and xpc−/−, either untreated or treated with UV irradiation at 5 hpi using TRIzol (Invitrogen). The mRNA library was constructed using the Illumina RNA library Prep Kit (NEB #E7775) and sequenced by Illumina HiSeq2500 (2 × 150 paired-end configuration).
Raw data in fastq format were processed by Cutadapt (V1.9.1, phred cutoff: 20, error rate: 0.1, adapter overlap: 1 bp, min. length: 75, proportion of N: 0.1) to be high quality clean data. The clean data were aligned to the reference genome (NCBI) using the software Hisat2 (v2.0.1). Gene expression was obtained using Htseq (v0.6.1). Differential expression analysis (DEA) was performed with the DESeq2 Bioconductor package in R[R Core Team (2022)]. The R statistical language and environment were used for principal component analysis (PCA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Among those differentially expressed genes between WT and xpc−/−, 11 genes were selected for the expression confirmation with qRT-PCR. The information on qRT-PCR primers is listed in Supplementary Table S2.
3. Results
3.1. Zebrafish xpc Gene
The zebrafish genome contains a single copy of the xpc gene located on chromosome 8. It is predicted that xpc gene has three isoforms: transcript 201, 202 and 203. Transcript 201 is the highest expressed isoform, which contains 14 exons and encodes Xpc full length protein with 879 amino acids (aa) [37]. Transcript 202, with an internal deletion of exon 7, encodes a protein of 844 aa without 275 to 305 aa in Xpc full length protein. Transcript 203 only contains the first eight exons and produces a protein of 238 aa without any known functional domains (Figure 1A and Figure S1A).
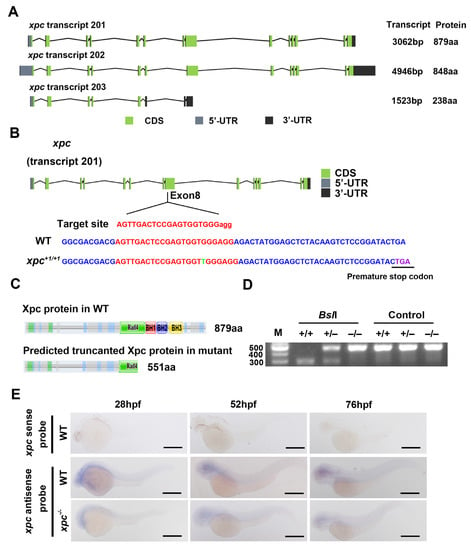
Figure 1.
Generation of zebrafish xpc−/− mutant. (A) Structural diagrams of the predicted three xpc isoforms in zebrafish. (B,C) Diagram showing the gRNA targeting site and one-bp insertion in the exon 8 of xpc mutant (B), which results in a premature stop codon (PTC) at 551 aa (C). Rad4, BH1, BH2 and BH3 are four known domains in Xpc protein. (D) Genotyping of xpc mutant with gel electrophoresis. A PCR product containing the mutation site was digested with BslI. M: DNA molecular weight marker; +/+: xpc WT; +/–: xpc heterozygotes; −/−: homozygotes. (E) Whole-mount in situ hybridization of the xpc gene with sense or antisense probes in WT and xpc−/− mutant embryos at 28, 52 and 76 hpf as indicated. Scale bar: 500 µm.
Protein alignments with other model organisms showed that the similarity of XPC protein between zebrafish (Transcript 201) and human, rat, mouse, yeast, or drosophila is 61.5%, 60.1%, 58.3%, 35.2%, or 32.4% respectively (Figure S1B). The phylogenetic tree showed that zebrafish Xpc protein is evolutionally closer to mammals than to drosophila and yeast (Figure S1C).
3.2. Generation of Zebrafish xpc−/− Mutant
To investigate the function of zebrafish xpc gene, we generated an xpc genetic mutant with the CRISPR/Cas9 technology. For this purpose, we designed a guide RNA (gRNA) to target the exon 8 of the xpc gene (Figure 1B) and co-injected the xpc-specific gRNA with the Cas9 protein into one-cell stage embryos. One allele was obtained from F1 progenies. This xpc mutant carries one-bp insertion in the exon 8, which results in a frameshift to the open reading frame (ORF) of xpc and introduces a premature stop codon (PTC) at the Rad4-domain. The predicted mutant protein lacks part of Rad4 domain and entire BH1, BH2, BH3 domains, suggesting that it is a dysfunctional protein (Figure 1C). The mutant fish was easily identified by a PCR fragment containing the mutation site. The gRNA target site in WT type contains a BslI recognition motif [CCN(7)GG], which was inserted with a T in the xpc mutant. Therefore, the PCR fragment from the wild type (WT) fish, but not from the mutant fish, was digested into two smaller fragments with a BslI restriction endonuclease (Figure 1D).
Similar to the xpc knockout mice, under non-stress conditions, the xpc−/− homozygous zebrafish develop relatively normally (without obvious phenotypes) and are fertile. To examine the spatial and temporal expression pattern of xpc during embryogenesis, we used antisense and sense probes of xpc to perform WISH assay. The positive signal was observed only from antisense probes, but not from sense probes, suggesting it was a true xpc expression signal. WISH showed that xpc expressed ubiquitously in whole embryos from 28 h post fertilization (hpf) to 76 hpf (Figure 1E). The signal intensity in xpc−/− mutant embryos decreased, compared to that in WT embryos. The results indicated that the level of the xpc mutant mRNA was lower than that of the WT mRNA.
3.3. Zebrafish xpc−/− Mutant Embryos Are More Sensitive to UV Irradiation
Studies from humans and mice have already shown that dysfunction of XPC results in increased photosensitivity and risk in skin associated diseases caused by UV irradiation. Therefore, we treated 28 hpf WT and xpc−/− embryos with UV irradiation at different doses of 75 J/m2 or 180 J/m2. As expected, the high dose of UV irradiation (180 J/m2) led to more severe abnormal development (such as edema, curve body and death) of both WT and mutant embryos at 72 h post treatment (hpt) than the low dose (75 J/m2) (Figure 2A). The percentage of abnormal embryos in xpc−/− mutants treated with 75 J/m2 or 180 J/m2 was 67.33% and 95.46% respectively, which was significantly higher than that in WT with corresponding treatments (34.67% for 75 J/m2 and 71.75% for 180 J/m2) (Figure 2B).
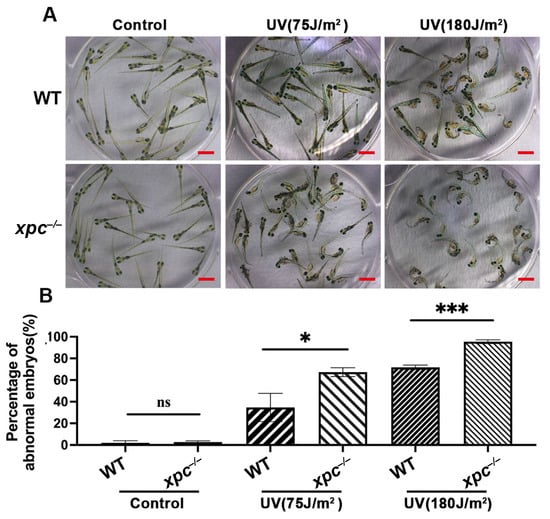
Figure 2.
UV irradiation causes more severe phenotypes in zebrafish xpc−/− mutant embryos. (A) Representative images of WT and xpc−/− embryos 100 h after UV irradiation. The 28 hpf WT and xpc−/− embryos were treated with UV irradiation at the dosage of either 0 (Control), 75 J/m2 or 180 J/m2. (B) Average percentage of abnormal embryos counted from image (A). There were at least 50 embryos in each treatment. Each experiment was repeated three times. Bars in graphs represent mean ± standard deviation. Two-tailed t-test was applied for each individual comparison (* p < 0.05, *** p < 0.001, n.s: no significance). Scale bar: 2 mm.
To investigate whether the increased abnormal development in the UV-treated xpc−/− embryos was related to the loss of the NER pathway, we performed comet assays to detect the accumulation of DNA damage in the WT and xpc−/− embryos treated or untreated with 180 J/m2 at 0 hpt and 24 hpt (Figure 3A). The results showed that the proportions of tail DNA contents were significantly increased in either UV-treated WT (42.11%) or UV-treated xpc−/− embryos (44.80%) at 0 hpt, compared to those in corresponding untreated controls (8.36% and 8.96% respectively) (Figure 3B). There was no significant difference in the proportions of tail DNA contents between untreated WT and untreated xpc−/− embryos, or between treated WT and treated xpc−/− embryos at 0 hpt. The results suggested that UV irradiation caused similar levels of DNA damages in both WT and xpc−/− embryos. At 24 hpt, the proportion of tail DNA contents in either treated WT or treated xpc−/− embryos was significantly decreased, compared to that in corresponding treated embryos at 0 hpt, suggesting that the DNA damage was being repaired. However, the proportion of tail DNA contents was significantly higher in UV-treated xpc−/− embryos (36.78%) than in UV-treated WT embryos (23.97%) (Figure 3B). The results demonstrated that the increased DNA damage accumulation in UV-treated xpc−/− embryos was due to loss-of-function of NER.
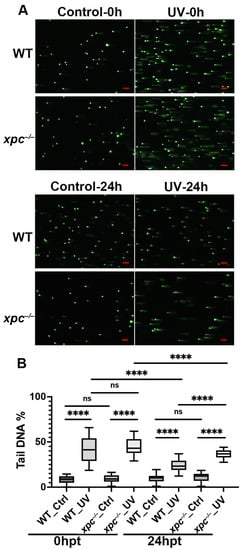
Figure 3.
Loss-of-function of Xpc leads to more DNA damage accumulation in UV-irradiated zebrafish embryos. (A) Representative images of comet assay in WT and xpc−/− embryos, 0 h and 24 h after UV irradiation. The 28 hpf WT and xpc−/− embryos were treated with UV irradiation at the dosage of either 0 (Control) or 180 J/m2. (B) Average percentage of tail DNA in each cell counted from image A. At least 50 cells were randomly selected in each microscopic field. Three microscopic fields were randomly taken in each treatment. Bars in graphs represent mean ± standard deviation. Two-tailed t-test was applied for each individual comparison (**** p < 0.0001, n.s: no significance). Scale bar: 100 µm.
Next, we performed a TUNEL assay to analyze whether the increased DNA damage led to apoptotic activity (Figure 4A). The results showed that there was no obvious difference in the intensity of TUNEL signals between untreated WT (33,934/embryo) and untreated xpc−/− embryos (43,841/embryo). However, at 24 hpt, the intensity of TUNEL signals was significantly higher in UV-treated xpc−/− embryos (5,691,695/embryo) than in UV-treated WT (1,194,601/embryo) (Figure 4B).
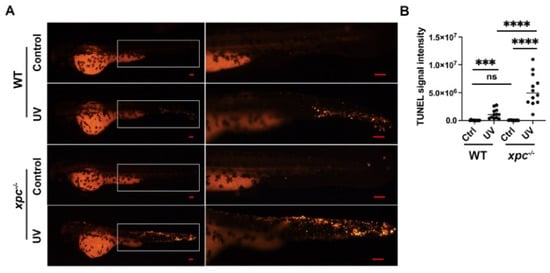
Figure 4.
Knockout of xpc results in more apoptotic activity in UV-irradiated zebrafish embryos. (A) Representative images of TUNEL assay in WT and xpc−/− embryos 24 h after UV irradiation. The 28 hpf WT and xpc−/− embryos were treated with UV irradiation at the dosage of either 0 (Control) or 75 J/m2. The area in the white rectangle was magnified in the right panel. (B) Average percentage TUNEL signal intensity in each embryo counted from image A. TUNEL signal intensity in each embryo was analyzed by imageJ software (TUNEL signal integrated density = TUNEL signal area*mean gray value). At least 15 embryos were randomly selected from each treatment. The experiment was repeated three times. Bars in graphs represent mean ± standard deviation. Two-tailed t-test was applied for each individual comparison (*** p < 0.001, **** p < 0.0001, n.s: no significance). Scale bar: 100 µm.
Taken together, the data demonstrated that the knockout of xpc sensitized zebrafish embryos to UV irradiation.
3.4. Transcriptomic Analysis Reveals That the Expression of p53 Signaling Pathway and Apoptotic Genes Is over Upregulated in xpc−/− Mutant Embryos upon UV Irradiaton
To explore if xpc plays a role in the regulation of transcription beyond its DNA damage repair function, we performed RNA-seq to analyze transcriptomes in untreated xpc−/− and WT embryos, as well as 180 J/m2 UV treated xpc−/− and WT embryos at 5 hpt. Each treatment had three independent replicates and the total RNA was extracted from 30 embryos in each replicate. Cleans reads from each of all twelve samples exceeded 5 GB, and the data filtering based on the Clean Q30 Bases Rate program showed that the Q30 for each of all twelve samples was more than 93%, demonstrating that the RNA-seq data obtained were of high quality. On average, 42,570,586 clean sequences (∼87% of total clean reads) matched a cDNA counterpart in the zebrafish genome (Danio rerio.GRCz11). The expression levels of zebrafish genes in each sample were calculated based on the method of fragments per kilobase millon mapped fragments (FPKM). Principal component analysis (PCA) was used to reduce the dimensionality of the dataset to visualize the global effect of UV irradiation and xpc mutation on the transcriptome. The four treatments were nicely separated in a 2D plot with different colors: red (xpc−/−_CK), green (WT_UV), blue (WT_CK) and orange(xpc−/−_UV) (Figure 5A). Clustering analysis using the hierarchical cluster also showed that the RNA-seq data were highly consistent among the three untreated WT, the three untreated mutant, the three UV-treated WT and the three UV-treated mutant samples (Figure 5B).
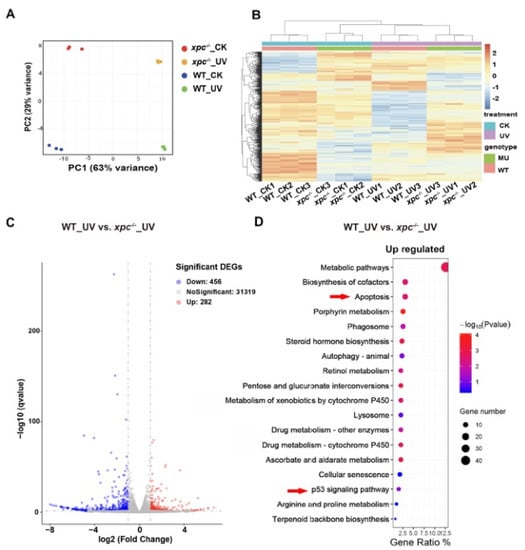
Figure 5.
Transcriptome analysis of WT and xpc−/− zebrafish embryos in response to UV irradiation. (A) Principal component analysis (PCA). Raw expression data from three independent replicates including untreated WT (WT_CK), UV-treated WT (WT_UV), untreated xpc−/− (xpc−/−_CK) and UV-treated xpc−/− mutant embryos (xpc−/−_UV), were subjected to principal component analysis (PCA). (B) Hierarchical clustering analysis of the RNA-seq data from WT_CK, WT_UV, xpc−/−_CK and xpc−/−_UV embryos, with three biological repeats. (C) Volcano plots showing the DEGs of WT_UV versus xpc−/−_UV. (D) Significantly enriched KEGG pathways for upregulated DEGs in xpc−/−_UV embryos compared with in WT_UV embryos.
A cross comparison of gene expression using the EdgeR method (V2.28.1) identified that 1423 genes were upregulated (log2 ≥ 1, p < 0.05) and 269 genes were downregulated (log2 ≤ –1, p < 0.05) in WT embryos upon UV irradiation (Figure S2A), whereas 239 genes were downregulated (log2 ≤ –1, p < 0.05) and 776 genes were upregulated (log2 ≥ 1, p < 0.05) in xpc−/− embryos upon UV irradiation (Figure S2B). Among the differentially expressed genes (DEGs) upon the UV irradiation, 573 genes were conserved between WT and xpc−/− (Figure S2C). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on the DEGs revealed that only two items (p53 signaling pathway and cell cycle) were enriched in the upregulated genes of treated WT embryos, which also appeared in the upregulated genes of treated xpc−/− embryos (Figure S2D,E). In addition to these two items, other nine signaling pathways (including apoptosis, protein processing in endoplasmic reticulum, MAPK signaling pathway, cellular senescence, herpes simplex virus 1 infection, FoxO signaling pathway, endocytosis and spliceosome) were also identified in the upregulated genes of treated xpc−/− embryos (Figure S2E).
To further analyze the differences between treated WT and treated xpc−/− mutant embryos, we compared transcriptomes between UV-treated WT and UV-treated xpc−/− embryos. A total of 456 genes were upregulated and 282 genes were downregulated in treated xpc−/− embryos (Figure 5C). KEGG analyses showed that the top item among the upregulated genes was metabolic pathways. Not surprisingly, apoptosis, cellular senescence and p53 signaling pathway were all enriched in upregulated genes (Figure 5D). The results suggested that DNA damage response upon UV irradiation triggered the p53 signaling pathway in both WT and xpc−/− embryos. Due to loss-of-function of NER, more DNA damage accumulated in treated xpc−/− embryos, which led to higher apoptosis, cellular senescence and other processes such as metabolic pathways, etc.
To explore if UV irradiation and knockout of xpc influenced the expression of genes in NER pathway, we searched 44 genes involved in NER pathway. Expectedly, the expression of xpc decreased about five times in either untreated or treated xpc−/− mutant embryos, compared to that in corresponding WT embryos (Figure S3A,B). The decrease in xpc expression in xpc−/− mutant embryos was confirmed by qRT-PCR (Figure S3C). These results were consistent with the observation in WISH experiment (Figure 1E). The results suggested that xpc mutant mRNA with a PTC was degraded by NMD pathway. To our surprise, only three genes (cetn2, rbx1 and ddb1) in WT and two genes (rbx1 and pcna) in xpc−/− mutant were upregulated (log2 ≥ 0.58), p < 0.05) by UV irradiation (Figure S3B). There was no significant difference in NER gene expression between treated WT and treated xpc−/− mutant. In normal conditions, only one gene (pold3, an accessory subunit of the replicative Pol δ polymerase) (log2 ≤ ™0.58, p < 0.05) was significantly downregulated in xpc−/− mutant. The results indicated that UV irradiation and knockout of xpc had little effect on the expression of genes in NER pathway.
3.5. A Total of 379 Differentially Expressed Genes in xpc−/− Mutant were Identified Not to Respond to UV Irradiation
Compared to untreated WT embryos, 972 genes including 710 upregulated genes (log2 ≥ 1, p < 0.05) and 262 downregulated genes (log2 ≤ –1, p < 0.05) were differentially expressed in untreated xpc−/− mutant embryos (Figure 6A). The cross comparison between these DEGs in untreated xpc−/− mutants and DEGs in treated xpc−/− mutants, or DEGs in treated WT embryos, revealed that among these 972 DEGs, the expression of 379 genes was not in response to UV irradiation in either WT or mutant embryos (Figure 6B, Table S1). To evaluate the RNA-seq data, we randomly selected 11 genes including one upregulated and 10 downregulated genes to perform qRT-PCR. The differential expression of these 11 genes was confirmed by qRT-PCR (Figure S4). The analysis indicates that the functions of Xpc also exceed the DNA damage response.
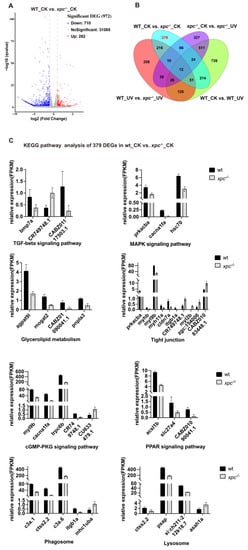
Figure 6.
The roles of Xpc beyond DNA damage repair. (A) Volcano plots showing the DEGs of WT_CK versus xpc−/−_CK. (B) Venn diagram showing the distribution of DEGs among four comparisons: WT_CK versus xpc−/−_CK, WT_CK versus WT_UV, xpc−/−_CK versus xpc−/−_UV, and WT_UV versus xpc−/−_UV. (C) KEGG pathway analysis of 379 DEGs in xpc−/−_CK compared with WT_CK, which did not respond to UV irradiation.
KEGG analysis of 379 DEGs showed that a number of signal pathways were enriched in the DEGs including TGF-β signaling (one gene upregulated and two genes downregulated), MAPK signaling (three genes downregulated), glycerolipid metabolism (four genes downregulated), tight junction (six genes downregulated and four genes upregulated), cGMP-PKG signaling (four genes downregulated and one gene upregulated), PPAR signaling (three genes downregulated), phagosome (four genes downregulated and one gene upregulated) and lysosome (three genes downregulated and one gene upregulated) (Figure 6C). By searching public databases, we found that among 379 DEGs, 86 genes (including 57 upregulated and 29 downregulated genes) are differentially expressed in cancer tissues (Table S1). The results suggested that in addition to its function in DNA damage repair, Xpc may also play a role in regulating the expression of some genes to contribute to tumorigenesis.
4. Discussion
It is well known that XPC plays an essential role in the recognition of bulky DNA lesions and subsequent activation of GG-NER [38]. Loss-of-function of XPC leads to development of UV-induced dermatologic malignancies and modifications of cancer response to chemotherapies including cisplatin [39]. There are also several lines of evidence to show that XPC defects are correlated not only with development of skin cancer but also with development of internal cancers such as lung, bladder, urinary, digestive organs, thyroid, breast, head, neck and leukemia [13]. Furthermore, recent studies have demonstrated that XPC plays a non-canonical role in other DNA repair mechanisms and tumor suppressor transcriptional regulations [39]. However, whether different functions of XPC play different roles in the development of internal cancers remain elusive.
Zebrafish provide several key advantages as a model system for studying human diseases. These include external fertilization, optical transparency, genome editing and easy high-throughput drug screens in live animals. Here, to further explore Xpc functions, we generated a zebrafish xpc loss-of-function mutant. Similar to the mouse model, no obvious phenotypes were observed in xpc−/− mutant under normal conditions. However, when exposed to UV irradiation, mutant embryos developed more severe abnormal phenotypes than the wild type embryos. Comet assay showed that irradiated xpc−/− mutant embryos accumulated significantly more unrepaired DNA damage than irradiated WT embryos at 24 hpt. TUNEL assay exhibited that significantly more apoptotic cells were observed in irradiated xpc−/− mutant embryos than in irradiated WT embryos. Furthermore, transcriptome analyses revealed that the p53 signal pathway and apoptosis were over upregulated in UV-irradiated mutant embryos, compared to the unirradiated WT embryos. The results demonstrate that the function of Xpc in NER is conserved in zebrafish. Loss-of-function of Xpc leads to accumulation of unrepaired DNA lesions, which activates the p53 signal pathway to trigger apoptotic activity in mutant embryos.
A total of 379 genes were identified to differentially express in xpc−/− mutant compared to those in WT embryos under normal conditions, and these genes did not respond to UV irradiation in either WT or xpc−/− mutant embryos. These DEGS were enriched in TGF-β signaling, MAPK signaling, glycerolipid metabolism, tight junction, cGMP-PKG signaling, PPAR signaling, phagosome and lysosome. The data indicate that Xpc may have a role in regulating these signaling pathways. However, whether these functions of Xpc other than DNA damage repair contribute to the development of internal cancers needs further investigation.
5. Conclusions
In summary, our results not only demonstrate that zebrafish Xpc has evolutionally conserved function in NER repair, but also show that Xpc has functions beyond DNA damage response. The zebrafish xpc−/− mutant will provide a platform to further investigate whether the function of Xpc beyond DNA repair is related to different internal cancers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes8040191/s1, Figure S1: Alignment of mRNA sequences of three xpc transcripts and amino acid sequence of six Xpc protein paralogues. Figure S2: Transcriptome analysis in UV treated WT and UV treated xpc−/− embryos compared to corresponding untreated controls. Figure S3: The influence of UV irradiation and xpc knockout on the expression of genes involved in NER pathway. Figure S4: The qPCR validation of RNA-Seq data. Supplement Table S1: List of primers used for qPCR validation. Supplement Table S2: List of the 379 DEGs in WT vs. xpc−/− of RNAseq.
Author Contributions
Conceptualization, K.L. and J.C.; Formal analysis, K.L.; Investigation, K.L.; Methodology, K.L.; Project administration, L.J.L. and J.C.; Resources, Z.S.; Software, C.Y.; Supervision, L.J.L.; Writing—original draft, K.L. and J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0801005), the National Natural Science Foundation of China (32192400 and 31871500), and the Starry Night Science Fund of Zhejiang University Shanghai Institute for Advanced Study (SN-ZJU-SIAS-004) for funding support.
Institutional Review Board Statement
All animal procedures were performed in full accordance with the requirements of the Regulation for the Use of Experimental Animals of Zhejiang Province. This work was specifically approved by the Animal Ethics Committee of the School of Medicine, Zhejiang University (ethics code permit no. ZJU20200071).
Informed Consent Statement
Not applicable.
Data Availability Statement
RNA-seq data has been deposited to the SRA database (accession number PRJNA895185). The data that support the findings of this report are available from the corresponding author upon request.The graphical abstract image was made by Figdraw.
Acknowledgments
We thank many people not listed as authors who provided help, encouragement and feedback.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Bowden, N.A. Nucleotide excision repair: Why is it not used to predict response to platinum-based chemotherapy? Cancer Lett. 2014, 346, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Nouspikel, T. DNA Repair in Mammalian Cells. Cell. Mol. Life Sci. 2009, 66, 994–1009. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Adar, S.; Selby, C.P.; Lieb, J.D.; Sancar, A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015, 29, 948–960. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.J.; Scharer, O.D. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006, 106, 253–276. [Google Scholar] [CrossRef] [PubMed]
- Sugasawa, K. Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair 2016, 44, 110–117. [Google Scholar] [CrossRef]
- Gavande, N.S.; VanderVere-Carozza, P.S.; Hinshaw, H.D.; Jalal, S.I.; Sears, C.R.; Pawelczak, K.S.; Turchi, J.J. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol. Ther. 2016, 160, 65–83. [Google Scholar] [CrossRef]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef]
- Fousteri, M.; Mullenders, L.H. Transcription-coupled nucleotide excision repair in mammalian cells: Molecular mechanisms and biological effects. Cell Res. 2008, 18, 73–84. [Google Scholar] [CrossRef]
- Renaud, E.; Miccoli, L.; Zacal, N.; Biard, D.S.; Craescu, C.T.; Rainbow, A.J.; Angulo, J.F. Differential contribution of XPC, RAD23A, RAD23B and CENTRIN 2 to the UV-response in human cells. DNA Repair 2011, 10, 835–847. [Google Scholar] [CrossRef]
- Sugasawa, K.; Masutani, C.; Uchida, A.; Maekawa, T.; Van Der Spek, P.J.; Bootsma, D.; Hoeijmakers, J.H.; Hanaoka, F. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol. 1996, 16, 4852–4861. [Google Scholar] [CrossRef]
- Okuda, Y.; Nishi, R.; Ng, J.M.; Vermeulen, W.; van der Horst, G.T.; Mori, T.; Hoeijmakers, J.H.J.; Hanaoka, F.; Sugasawa, K. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair 2004, 3, 1285–1295. [Google Scholar] [CrossRef]
- Ng, J.M.Y.; Vermeulen, W.; van der Horst, G.T.; Bergink, S.; Sugasawa, K.; Vrieling, H.; Hoeijmakers, J.H. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003, 17, 1630–1645. [Google Scholar] [CrossRef]
- Zebian, A.; Shaito, A.; Mazurier, F.; Rezvani, H.R.; Zibara, K. XPC beyond nucleotide excision repair and skin cancers. Mutat. Res.-Rev. Mutat. Res. 2019, 782, 108286. [Google Scholar] [CrossRef]
- Apelt, K.; Lans, H.; Schärer, O.D.; Luijsterburg, M.S. Nucleotide excision repair leaves a mark on chromatin: DNA damage detection in nucleosomes. Cell. Mol. Life Sci. 2021, 78, 7925–7942. [Google Scholar] [CrossRef]
- Lehmann, A.R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 2003, 85, 1101–1111. [Google Scholar]
- Bradford, P.T.; Goldstein, A.M.; Tamura, D.; Khan, S.G.; Ueda, T.; Boyle, J.; Oh, K.-S.; Imoto, K.; Inui, H.; Moriwaki, S.-I.; et al. Cancer and neurologic degeneration in xeroderma pigmentosum: Long term follow-up characterises the role of DNA repair. J. Med. Genet. 2011, 48, 168–176. [Google Scholar] [CrossRef]
- Kraemer, K.H.; Lee, M.M.; Scotto, J. Xeroderma-Pigmentosum—Cutaneous, Ocular, and Neurologic Abnormalities in 830 Published Cases. Arch. Dermatol. 1987, 123, 241–250. [Google Scholar] [CrossRef]
- Sands, A.T.; Abuin, A.; Sanchez, A.; Conti, C.J.; Bradley, A. High Susceptibility to Ultraviolet-Induced Carcinogenesis in Mice Lacking Xpc. Nature 1995, 377, 162–165. [Google Scholar]
- Hollander, M.C.; Philburn, R.T.; Patterson, A.D.; Velasco-Miguel, S.; Friedberg, E.C.; Linnoila, R.I.; Fornace, A.J., Jr. Deletion of XPC leads to lung tumors in mice and is associated with early events in human lung carcinogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 13200–13205. [Google Scholar]
- Chen, Z.W.; Yang, J.; Wang, G.; Song, B.; Li, J.; Xu, Z. Attenuated expression of xeroderma pigmentosurn group C is associated with critical events in human bladder cancer carcinogenesis and progression. Cancer Res. 2007, 67, 4578–4585. [Google Scholar] [CrossRef]
- Cheo, D.L.; Burns, D.K.; Meira, L.B.; Houle, J.F.; Friedberg, E.C. Mutational inactivation of the xeroderma pigmentosum group C gene confers predisposition to 2-acetylaminofluorene-induced liver and lung cancer and to spontaneous testicular cancer in Trp53(−/−) mice. Cancer Res. 1999, 59, 771–775. [Google Scholar] [PubMed]
- Meira, L.B.; Reis, A.M.; Cheo, D.L.; Nahari, D.; Burns, D.K.; Friedberg, E.C. Cancer predisposition in mutant mice defective in multiple genetic pathways: Uncovering important genetic interactions. Mutat. Res.-Fundam. Mol. Mech. Mutagen. 2001, 477, 51–58. [Google Scholar] [CrossRef]
- Nemzow, L.; Lubin, A.; Zhang, L.; Gong, F. XPC: Going where no DNA damage sensor has gone before. DNA Repair 2015, 36, 19–27. [Google Scholar] [CrossRef]
- Le May, N.; Mota-Fernandes, D.; Vélez-Cruz, R.; Iltis, I.; Biard, D.; Egly, J.M. NER Factors Are Recruited to Active Promoters and Facilitate Chromatin Modification for Transcription in the Absence of Exogenous Genotoxic Attack. Mol. Cell 2010, 38, 54–66. [Google Scholar] [CrossRef]
- Ho, J.J.; Cattoglio, C.; McSwiggen, D.T.; Tjian, R.; Fong, Y.W. Regulation of DNA demethylation by the XPC DNA repair complex in somatic and pluripotent stem cells. Genes Dev. 2017, 31, 830–844. [Google Scholar] [CrossRef]
- Bidon, B.; Iltis, I.; Semer, M.; Nagy, Z.; Larnicol, A.; Cribier, A.; Benkirane, M.; Coin, F.; Egly, J.-M.; Le May, N. XPC is an RNA polymerase II cofactor recruiting ATAC to promoters by interacting with E2F1. Nat. Commun. 2018, 9, 2610. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Xiao, A.; Wang, Z.; Hu, Y.; Wu, Y.; Luo, Z.; Yang, Z.; Zu, Y.; Li, W.; Huang, P.; Tong, X.; et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013, 41, e141. [Google Scholar] [CrossRef]
- Labun, K.; Montague, T.G.; Krause, M.; Torres Cleuren, Y.N.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019, 47, W171–W174. [Google Scholar] [CrossRef]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Ahkmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; Schier, A.F. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLoS ONE 2014, 9, e106396. [Google Scholar] [CrossRef]
- Zhao, S.Y.; Chen, Y.; Chen, F.; Huang, D.; Shi, H.; Lo, L.J.; Chen, J.; Peng, J. Sas10 controls ribosome biogenesis by stabilizing Mpp10 and delivering the Mpp10-Imp3-Imp4 complex to nucleolus. Nucleic Acids Res. 2019, 47, 2996–3012. [Google Scholar] [CrossRef]
- Gong, L.; Gong, H.; Pan, X.; Chang, C.; Ou, Z.; Ye, S.; Yin, L.; Yang, L.; Tao, T.; Zhang, Z.; et al. p53 isoform Delta 113p53/Delta 133p53 promotes DNA double-strand break repair to protect cell from death and senescence in response to DNA damage. Cell Res. 2015, 25, 351–369. [Google Scholar] [CrossRef]
- Azqueta, A.; Collins, A.R. The essential comet assay: A comprehensive guide to measuring DNA damage and repair. Arch. Toxicol. 2013, 87, 949–968. [Google Scholar] [CrossRef]
- Chen, J.; Ruan, H.; Ng, S.M.; Gao, C.; Soo, H.M.; Wu, W.; Zhang, Z.; Wen, Z.; Lane, D.P.; Peng, J.; et al. Loss of function of def selectively up-regulates Delta 113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005, 19, 2900–2911. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef]
- Nasrallah, N.A.; Wiese, B.M.; Sears, C.R. Xeroderma Pigmentosum Complementation Group C (XPC): Emerging Roles in Non-Dermatologic Malignancies. Front. Oncol. 2022, 12, 846965. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).