The Complete Mitochondrial Genome of the Fivespot Flounder, Pseudorhombus pentophthalmus (Pleuronectiformes: Paralichthyidae), from Korea and Its Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample and DNA Extraction
2.2. Illumina Sequencing, and Mitogenome Assembly and Annotation
2.3. Sequence Analysis
2.4. Phylogenetic Analysis
3. Results and Discussion
3.1. Mitogenome Organization of Pseudorhombus Pentophthalmus
3.2. Protein-Coding Genes (PCGs)
3.3. Ribosomal RNA and Transfer RNA Genes
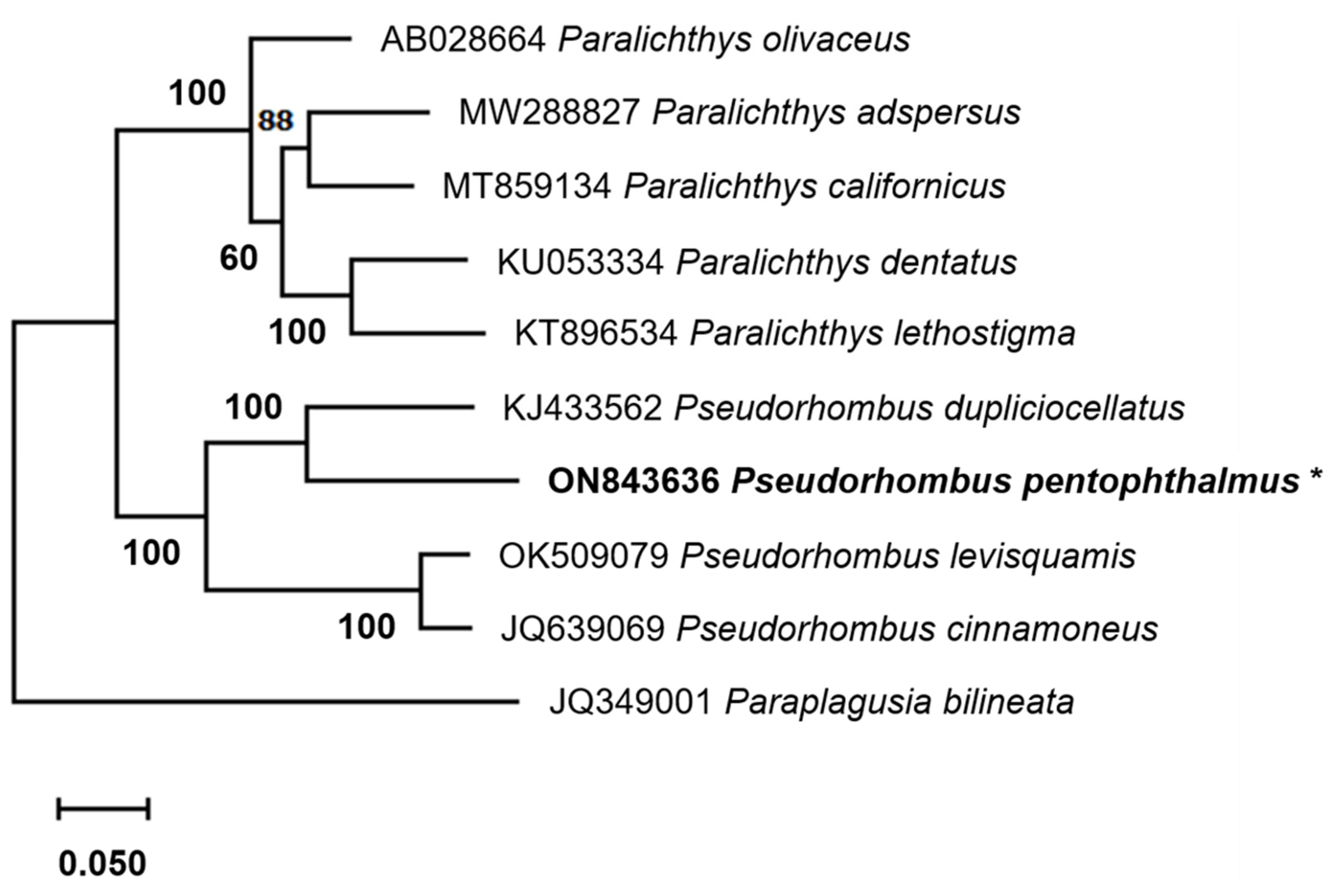
3.4. Phylogenetic Relationships
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, T.W. Seasonal variation in species composition of demersal fish in Yongil Bay, east coast of Korea. J. Korean Fish Soc. 1999, 32, 512–519. [Google Scholar]
- Park, J.M. Species Composition and Reproductive Ecology of Fishes in the Coastal Waters off Gori, Korea. Ph.D. Thesis, Pukyong National University, Busan, Republic of Korea, 2010. [Google Scholar]
- FAO. Food and Agriculture Organization of the United Nations. 2020. Available online: http://www.fao.org/fishery/species/3350/en (accessed on 25 January 2023).
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods 2008, 5, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Chinmayee, C.; Nischal, A.; Manjunath, C.R.; Soumya, K.N. Next Generation Sequencing in Big Data. Int. J. Trend Sci. Res. Dev. 2018, 2, 379–389. [Google Scholar]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Harrison, R.G. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trend Ecol. Evol. 1989, 4, 6–11. [Google Scholar] [CrossRef]
- Berendzen, P.B.; Dimmick, W.W. Phylogenetic relationships of Pleuronectiformes based on molecular evidence. Copeia 2002, 2002, 642–652. [Google Scholar] [CrossRef]
- Zhang, J.B.; Hanner, R. DNA barcoding is a useful tool for the identification of marine fishes from Japan. Biochem. System Ecol. 2011, 39, 31–42. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data (Babraham Bioinformatics, 2010). Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 3 February 2023).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide compositin at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–359. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Si, L.Z.; Gong, L.; Shi, W.; Yang, M.; Kong, X.Y. The complete mitochondrial genome of Pseudorhombus dupliocellatus (Pleuronectiformes: Paralichthyidae). Mitochondrial DNA Part A 2017, 28, 58–59. [Google Scholar] [CrossRef]
- Vargas-Peralta, C.E.; Farfán, C.; Barón-Sevilla, B.; Río-Portilla, M.A.D. Complete mitochondrial genome of the California halibut, Paralichthys californicus. Cienc. Mar. 2020, 46, 297–306. [Google Scholar] [CrossRef]
- Marín, A.; López-Landavery, E.; González-Martinez, S.; Reyes-Flores, L.E.; Corona-Herrera, G.; Tapia-Morales, S.; Yzásiga-Barrera, C.G.; Fernandino, J.I.; Zelada-Mázmela, E. The complete mitochondrial genome of the fine flounder Paralichthys adspersus revealed by next-generation sequencing. Mitochondrial DNA Part B Resour. 2021, 6, 2785–2787. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Liu, X.; Xu, Y.; Wang, B. The complete mitochondrial genome of southern flounder Paralichthys lethostigma (Pleuronectiformes, Bothidae). Mitochondrial DNA Part B Resour. 2016, 1, 200–201. [Google Scholar] [CrossRef]
- Shi, W.; Dong, X.L.; Wang, Z.M.; Miao, X.G.; Wang, S.Y.; Kong, X.Y. Complete mitogenome sequences of four flatfishes (Pleuronectiformes) reveal a novel gene arrangement of L-strand coding genes. BMC Evol. Biol. 2013, 13, 173. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, K.; Hayashizaki, K.; Yokoyama, Y.; Asahida, T.; Toyohara, H.; Yamashita, Y. Complete nucleotide sequence of Japanese flounder (Paralichthys olivaceus) mitochondrial genome: Structural properties and cue for resolving teleostean relationship. J. Hered. 2000, 91, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, X.; Shi, B.; Wang, B. Complete mitochondrial genome of summer flounder Paralichthys dentatus (Pleuronectiformes, Paralichthyidae). Mitochondrial DNA Part B Resour. 2016, 1, 889–890. [Google Scholar] [CrossRef]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]

| Species Studied | GenBank (BlastN) | Reference | |||
|---|---|---|---|---|---|
| Species Identified | GenBank no. | Length (bp) | Similarity (%) | ||
| Pseudorhombus pentophthalmus (Accession no.: ON843636; Length: 16,684 bp) | Pseudorhombus dupliciocellatus | KJ433562 | 16,621 | 83.06 | [22] |
| Paralichthys californicus | MT859134 | 16,858 | 79.30 | [23] | |
| Paralichthys adspersus | MW288827 | 17,060 | 78.80 | [24] | |
| Pseudorhombus levisquamis | OK509079 | 16,604 | 78.74 | NA | |
| Paralichthys lethostigma | KT896534 | 16,843 | 78.71 | [25] | |
| Pseudorhombus cinnamoneus | JQ639069 | 16,599 | 78.58 | [26] | |
| Paralichthys olivaceus | AB028664 | 17,090 | 78.54 | [27] | |
| Paralichthys dentatus | KU053334 | 17,033 | 78.18 | [28] | |
| Paraplagusia bilineata | JQ349001 | 16,985 | 75.15 | NA | |
| Gene | Strand | Start | End | Size (bp) | Start Codon | Stop Codon | Intergenic Nucleotides * |
|---|---|---|---|---|---|---|---|
| tRNA-Phe | H | 1 | 69 | 69 | - | - | - |
| 12SrRNA | H | 70 | 1011 | 942 | - | - | - |
| tRNA-Val | H | 1012 | 1084 | 73 | - | - | - |
| 16SrRNA | H | 1085 | 2811 | 1727 | - | - | - |
| tRNA-Leu | H | 2812 | 2885 | 74 | - | - | - |
| ND1 | H | 2886 | 3860 | 975 | ATA | TAG | 3 |
| tRNA-Ile | H | 3864 | 3934 | 71 | - | - | −1 |
| tRNA-Gln | L | 3934 | 4004 | 71 | - | - | −1 |
| tRNA-Met | H | 4004 | 4072 | 69 | - | - | - |
| ND2 | H | 4073 | 5117 | 1045 | GTG | T-- | - |
| tRNA-Trp | H | 5118 | 5190 | 73 | - | - | 1 |
| tRNA-Ala | L | 5192 | 5260 | 69 | - | - | 1 |
| tRNA-Asn | L | 5262 | 5334 | 73 | - | - | 37 |
| tRNA-Cys | L | 5372 | 5436 | 65 | - | - | - |
| tRNA-Tyr | L | 5437 | 5503 | 67 | - | - | 1 |
| COX1 | H | 5505 | 7049 | 1545 | GTG | TAG | 3 |
| tRNA-Ser | L | 7053 | 7123 | 71 | - | - | 9 |
| tRNA-Asp | H | 7133 | 7203 | 71 | - | - | 7 |
| COX2 | H | 7211 | 7901 | 691 | ATG | T-- | - |
| tRNA-Lys | H | 7902 | 7974 | 73 | - | - | 1 |
| ATP8 | H | 7976 | 8143 | 168 | ATG | TAA | −10 |
| ATP6 | H | 8134 | 8816 | 683 | ATG | TA- | - |
| COX3 | H | 8817 | 9601 | 785 | ATG | TA- | - |
| tRNA-Gly | H | 9602 | 9673 | 72 | - | - | - |
| ND3 | H | 9674 | 10022 | 349 | GTG | T-- | - |
| tRNA-Arg | H | 10023 | 10091 | 69 | - | - | - |
| ND4L | H | 10092 | 10388 | 297 | ATG | TAA | −7 |
| ND4 | H | 10382 | 11762 | 1381 | ATG | T-- | - |
| tRNA-His | H | 11763 | 11832 | 70 | - | - | - |
| tRNA-Ser | H | 11833 | 11899 | 67 | - | - | 4 |
| tRNA-Leu | H | 11904 | 11976 | 73 | - | - | 2 |
| ND5 | H | 11979 | 13817 | 1839 | ATG | TAA | −4 |
| ND6 | L | 13814 | 14335 | 522 | ATG | TAG | - |
| tRNA-Glu | L | 14336 | 14404 | 69 | - | - | 2 |
| CytB | H | 14407 | 15546 | 1140 | ATG | TAA | 1 |
| tRNA-Thr | H | 15548 | 15619 | 72 | - | - | 1 |
| tRNA-Pro | L | 15621 | 15691 | 71 | - | - | - |
| D-loop | H | 15692 | 16684 | 993 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-S.; Patil, M.P.; Kim, J.-O.; Lee, Y.-J.; Seo, Y.B.; Kim, J.-K.; Suryawanshi, R.K.; Kim, G.-D. The Complete Mitochondrial Genome of the Fivespot Flounder, Pseudorhombus pentophthalmus (Pleuronectiformes: Paralichthyidae), from Korea and Its Phylogenetic Analysis. Fishes 2023, 8, 150. https://doi.org/10.3390/fishes8030150
Lee Y-S, Patil MP, Kim J-O, Lee Y-J, Seo YB, Kim J-K, Suryawanshi RK, Kim G-D. The Complete Mitochondrial Genome of the Fivespot Flounder, Pseudorhombus pentophthalmus (Pleuronectiformes: Paralichthyidae), from Korea and Its Phylogenetic Analysis. Fishes. 2023; 8(3):150. https://doi.org/10.3390/fishes8030150
Chicago/Turabian StyleLee, Yong-Suk, Maheshkumar Prakash Patil, Jong-Oh Kim, Yu-Jin Lee, Yong Bae Seo, Jin-Koo Kim, Rahul K. Suryawanshi, and Gun-Do Kim. 2023. "The Complete Mitochondrial Genome of the Fivespot Flounder, Pseudorhombus pentophthalmus (Pleuronectiformes: Paralichthyidae), from Korea and Its Phylogenetic Analysis" Fishes 8, no. 3: 150. https://doi.org/10.3390/fishes8030150
APA StyleLee, Y.-S., Patil, M. P., Kim, J.-O., Lee, Y.-J., Seo, Y. B., Kim, J.-K., Suryawanshi, R. K., & Kim, G.-D. (2023). The Complete Mitochondrial Genome of the Fivespot Flounder, Pseudorhombus pentophthalmus (Pleuronectiformes: Paralichthyidae), from Korea and Its Phylogenetic Analysis. Fishes, 8(3), 150. https://doi.org/10.3390/fishes8030150








