Genetic Connectivity of Roundjaw Bonefish Albula glossodonta (Elopomorpha, Albulidae) in the Central Pacific Ocean Resolved through ddRAD-Based Population Genomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Collection Methods
2.2. Laboratory Methods
2.3. Data Analysis
3. Results
3.1. GSI Assays
3.2. Genomic Libraries
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hildebrand, S.F. Family Albulidae. In Fishes of the Western North Atlantic; Bigelow, H.B., Ed.; Sears Foundation for Marine Research, Bingham Oceanographic Laboratory, Yale University: New Haven, CT, USA, 1963; pp. 132–147. [Google Scholar]
- Bowen, B.; Karl, S.A.; Pfeiler, E. Resolving evolutionary lineages and taxonomy of bonefishes (Albula spp.). In Biology and Management of the World Tarpon and Bonefish Fisheries; Ault, J.S., Ed.; CRC Press: Boca Raton, FL, USA, 2008; pp. 147–154. [Google Scholar]
- Hidaka, K.; Iwatsuki, Y.; Randall, J.E. A review of the Indo-Pacific bonefishes of the Albula argentea complex, with a description of a new species. Ichthyol. Res. 2008, 55, 53–64. [Google Scholar] [CrossRef]
- Wallace, E.M.; Tringali, M.D. Identification of a novel member in the family Albulidae (bonefishes). J. Fish Biol. 2010, 76, 1972–1983. [Google Scholar] [CrossRef] [PubMed]
- Wallace, E.M. High intraspecific genetic connectivity in the Indo-Pacific bonefishes: Implications for conservation and management. Environ. Biol. Fishes 2015, 98, 2173–2186. [Google Scholar] [CrossRef]
- Wallace, E.M.; Tringali, M.D. Fishery composition and evidence of population structure and hybridization in the Atlantic bonefish species complex (Albula spp.). Mar. Biol. 2016, 163, 142. [Google Scholar] [CrossRef]
- Adams, A.J.; Cooke, S.J. Advancing the science and management of flats fisheries for bonefish, tarpon, and permit. Environ. Biol. Fishes 2015, 98, 2123–2131. [Google Scholar] [CrossRef]
- Donovan, M.K.; Friedlander, A.M.; Harding, K.K.; Schemmel, E.M.; Filous, A.; Kamikawa, K.; Torkelson, N. Ecology and niche specialization of two bonefish species in Hawai‘i. Environ. Biol. Fishes 2015, 98, 2159–2171. [Google Scholar] [CrossRef]
- Kamikawa, K.T.; Friedlander, A.M.; Harding, K.K.; Filous, A.; Donovan, M.K.; Schemmel, E. Bonefishes in Hawai‘i and the importance of angler-based data to inform fisheries management. Environ. Biol. Fishes 2015, 98, 2147–2157. [Google Scholar] [CrossRef]
- Filous, A.; Lennox, R.J.; Coleman, R.R.; Friedlander, A.M.; Clua, E.E.G.; Danylchuk, A.J. Life-history characteristics of an exploited bonefish Albula glossodonta population in a remote South Pacific atoll. J. Fish Biol. 2019, 95, 562–574. [Google Scholar] [CrossRef]
- Filous, A.; Lennox, R.J.; Eveson, P.; Raveino, R.; Clua, E.E.G.; Cooke, S.J.; Danylchuk, A.J. Population dynamics of roundjaw bonefish Albula glossodonta at a remote coralline Atoll inform community-based management in an artisanal fishery. Fish. Manag. Ecol. 2019, 27, 200–214. [Google Scholar] [CrossRef]
- Shaklee, J.B.; Tamaru, C.S. Biochemical and morphological evolution of Hawaiian bonefishes (Albula). Syst. Zool. 1981, 30, 125–146. [Google Scholar] [CrossRef]
- Pickett, B.D.; Wallace, E.M.; Ridge, P.G.; Kauwe, J.S.K. Lingering Taxonomic Challenges Hinder Conservation and Management of Global Bonefishes. Fisheries 2020, 45, 347–358. [Google Scholar] [CrossRef]
- Allen, M. The historical role of bonefishes (Albula spp.) in Polynesian fisheries. Hawaii. Archaeol. 2014, 4, 51–72. [Google Scholar]
- Adams, A.; Guindon, K.; Horodysky, A.; MacDonald, T.; McBride, R.; Shenker, J.; Ward, R. Albula glossodonta. The IUCN Red List of Threatened Species. Retrieved Oct. 2012, 2012, e.T194299A2310398. [Google Scholar]
- Adams, A.; Guindon, K.; Horodysky, A.; MacDonald, T.; McBride, R.; Shenker, J.; Ward, R. Albula virgata. The IUCN Red List of Threatened Species. Retrieved Oct. 2012, 2012, e.T194302A2310633. [Google Scholar]
- Gaither, M.R.; Jones, S.A.; Kelley, C.; Newman, S.J.; Sorenson, L.; Bowen, B.W. High Connectivity in the Deepwater Snapper Pristipomoides filamentosus (Lutjanidae) across the Indo-Pacific with Isolation of the Hawaiian Archipelago. PLoS ONE 2011, 6, e28913. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.E.; McDowell, J.R. Population structure of istiophorid billfishes. Fish. Res. 2015, 166, 21–28. [Google Scholar] [CrossRef]
- Friedlander, A.M.; Caselle, J.E.; Beets, J.; Lowe, C.G.; Bowen, B.W.; Ogawa, T.K.; Kelley, K.M.; Calitri, T.; Lange, M.; Anderson, B.S. Biology and ecology of the recreational bonefish fishery at Palmyra Atoll National Wildlife Refuge with comparisons to other Pacific Islands. In Biology and Management of the World Tarpon and Bonefish Fisheries; Ault, J.S., Ed.; CRC Press: Boca Raton, FL, USA, 2008; pp. 28–56. [Google Scholar]
- Hourigan, T.F.; Reese, E.S. Mid-ocean isolation and the evolution of Hawaiian reef fishes. Trends Ecol. Evol. 1987, 2, 187–191. [Google Scholar] [CrossRef]
- Craig, M.T.; Eble, J.; Bowen, B.W. Origins, ages, and populations histories: Comparative phylogeography of endemic Hawaiian butterfly fishes (genus Chaetodon). J. Biogeogr. 2010, 37, 2125–2136. [Google Scholar] [CrossRef]
- Hodge, J.R.; van Herwerden, L.; Bellwood, D.R. Temporal evolution of coral reef fishes: Global patterns and disparity in isolated locations. J. Biogeogr. 2014, 41, 2115–2127. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Gaither, M.R.; Bernal, M.A.; Coleman, R.R.; Bowen, B.W.; Jones, S.A.; Simison, W.B.; Rocha, L.A. Genomic signatures of geographic isolation and natural selection in coral reef fishes. Mol. Ecol. 2015, 24, 1543–1557. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.J.; Horodysky, A.Z.; McBride, R.S.; Guindon, K.; Shenker, J.; MacDonald, T.C.; Harwell, H.D.; Ward, R.; Carpenter, K. Global conservation status and research needs for tarpons (Megalopidae), ladyfishes (Elopidae) and bonefishes (Albulidae). Fish Fish. 2013, 15, 280–311. [Google Scholar] [CrossRef]
- Wilson, J.K.; Adams, A.J.; Ahrens, R.N.M. Atlantic tarpon (Megalops atlanticus) nursery habitats: Evaluation of habitat quality and broad-scale habitat identification. Environ. Biol. Fishes 2019, 102, 383–402. [Google Scholar] [CrossRef]
- Fernandez, C.A.F.; Cunha, F.E.A.; Silva, C.E.L.S.; Araujo, A.C.S.; Pereira, R.L.; Viana, D.F.; Magalhães, W.M.; Gondolo, M.A.P.; de Castro, D.M.; Adams, A.; et al. Population dynamics and movements of Atlantic tarpon, Megalops atlanticus, in the Paranaiba Delta Protected Area, Brazil: Challenges for local fishery management planning. Environ. Biol. Fishes 2022, 106, 449–468. [Google Scholar] [CrossRef]
- Yochum, N.; Starr, R.M.; Wendt, D.E. Utilizing Fishermen Knowledge and Expertise: Keys to Success for Collaborative Fisheries Research. Fisheries 2011, 36, 593–605. [Google Scholar] [CrossRef]
- Schemmel, E.; Friedlander, A.M.; Andrade, P.; Keakealani, K.; Castro, L.M.; Wiggins, C.; Wilcox, B.A.; Yasutake, Y.; Kittinger, J.N. The codevelopment of coastal fisheries monitoring methods to support local management. Ecol. Soc. 2016, 21, 34. [Google Scholar] [CrossRef]
- Seyoum, S.; Wallace, E.M.; Tringali, M.D. Permanent genetic resources: Twelve polymorphic microsatellite markers for the bonefish, Albula vulpes and two congeners. Mol. Ecol. Resour. 2008, 8, 354–356. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX, Logiciel sous WindowsTM pour la Génétique des Populations; Laboratoire Génome, Populations, Interactions CNRS UMR 5000, Université deMontpellier II: Montpellier, France, 2000. [Google Scholar]
- Faircloth, B.; Glenn, T. Homemade AMPure XP Beads; Department of Ecology and Evolutionary Biology, University of California: Los Angeles, CA, USA, 2011. [Google Scholar]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- McKinney, G.J.; Waples, R.K.; Seeb, L.W.; Seeb, J.E. Paralogs are revealed by proportion of heterozygotes and deviations in read ratios in genotyping-by-sequencing data from natural populations. Mol. Ecol. Resour. 2016, 17, 656–669. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Green, P.; Kamvar, Z.; Gosselin, T. mmod: Modern Measures of Population Differentiation (Version 1.3.3). 2017. Available online: https://github.com/dwinter/mmod (accessed on 1 October 2023).
- Hedrick, P.H. Genetics of Populations, 4th ed.; James and Bartlett Publishers: Subury, MA, USA, 2011. [Google Scholar]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmend, I. Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational Inference of Population Structure in Large SNP Data Sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Toonen, R.J.; Andrews, K.R.; Baums, I.B.; Bird, C.E.; Concepcion, G.T.; Daly-Engel, T.S.; Eble, J.A.; Faucci, A.; Gaither, M.R.; Iacchei, M.; et al. Defining Boundaries for Ecosystem-Based Management: A Multispecies Case Study of Marine Connectivity across the Hawaiian Archipelago. J. Mar. Biol. 2011, 2011, 460173. [Google Scholar] [CrossRef]
- Lindo-Atichati, D.; Jia, Y.; Wren, J.L.K.; Antoniades, A.; Kobayashi, D.R. Eddies in the Hawaiian Archipelago Region: Formation, Characterization, and Potential Implications on Larval Retention of Reef Fish. J. Geophys. Res. Oceans 2020, 125, e2019JC015348. [Google Scholar] [CrossRef]
- Rivera, M.A.J.; Andrews, K.R.; Kobayashi, D.R.; Wren, J.L.K.; Kelley, C.; Roderick, G.K.; Toonen, R.J. Genetic Analyses and Simulations of Larval Dispersal Reveal Distinct Populations and Directional Connectivity across the Range of the Hawaiian Grouper (Epinephelus quernus). J. Mar. Biol. 2010, 2011, 765353. [Google Scholar] [CrossRef]
- Tenggardjaja, K.A.; Bowen, B.W.; Bernardi, G. Reef Fish Dispersal in the Hawaiian Archipelago: Comparative Phylogeography of Three Endemic Damselfishes. J. Mar. Biol. 2016, 2016, 3251814. [Google Scholar] [CrossRef]
- Coleman, R.R.; Bowen, B.W. Genomic assessment of an endemic Hawaiian surgeonfish, Acanthurus triostegus sandvicensis reveals high levels of connectivity and fine-scale population structure. Coral Reefs 2022, 41, 687–697. [Google Scholar] [CrossRef]
- Rivera, M.A.J.; Kelley, C.D.; Roderick, G.K. Subtle population genetic structure in the Hawaiian grouper, Epinephelus quernus (Serranidae) as revealed by mitochondrial DNA analyses. Biol. J. Linn. Soc. 2004, 81, 449–468. [Google Scholar] [CrossRef]
- Williams, C.T.; Mclvor, A.J.; Wallace, E.M.; Lin, Y.J.; Berumen, M.L. Genetic diversity and life-history traits of bonefish Albula spp. from the Red Sea. J. Fish Biol. 2020, 98, 855–864. [Google Scholar] [CrossRef]
- Husson, L.; Boucher, F.C.; Sarr, A.; Sepulchre, P.; Cahyarini, S.Y. Evidence of Sundaland’s subsidence requires revisiting its biogeography. J. Biogeogr. 2019, 47, 843–853. [Google Scholar] [CrossRef]
- Danylchuk, A.J.; Cooke, S.J.; Goldberg, T.L.; Suski, C.D.; Murchie, K.J.; Danylchuk, S.E.; Shultz, A.D.; Haak, C.R.; Brooks, J.; Oronti, A.; et al. Aggregations and offshore movements as indicators of spawning activity of bonefish (Albula vulpes) in The Bahamas. Mar. Biol. 2011, 158, 1981–1999. [Google Scholar] [CrossRef]
- Wills, P.S.; Shenker, J.; Ajemian, M.J.; Mejri, S.; Adams, A. Bonefish Reproduction Research Project: March 2016–February 2022 Final Report. 2022, p. 189. Available online: https://www.bonefishtarpontrust.org/downloads/research-reports/stories/brrp-btt-final%20report.pdf (accessed on 11 September 2022).
- Lombardo, S.M.; Chérubin, L.M.; Adams, A.J.; Shenker, J.M.; Wills, P.S.; Danylchuk, A.J.; Ajemian, M.J. Biophysical larval dispersal models of observed bonefish (Albula vulpes) spawning events in Abaco, The Bahamas: An assessment of population connectivity and ocean dynamics. PLoS ONE 2022, 17, e0276528. [Google Scholar] [CrossRef]
- Talma, S.C.A.L. Genetic Connectivity of the Roundjaw Bonefish (Albula glossodonta) in the Southwest Indian Ocean. Master’s Thesis, Rhodes University, Grahamstown, South Africa, 2021. Available online: http://hdl.handle.net/10962/192174 (accessed on 10 February 2023).
- Douglas, M.R.; Chafin, T.K.; Claussen, J.E.; Philipp, D.P.; Douglas, M.E. Are populations of economically important bonefish and queen conch ‘open’ or ‘closed’ in the northern caribbean basin? Mar. Ecol. 2021, 42, e12639. [Google Scholar] [CrossRef]
- Maragos, J.E.; Jokiel, P.L. Reef corals of Johnston Atoll: One of the world’s most isolated reefs. Coral Reefs 1986, 4, 141–150. [Google Scholar] [CrossRef]
- Kosaki, R.K.; Pyle, R.L.; Randall, J.E.; Irons, D.K. New records of fishes from Johnston Atoll with notes on biogeography. Pac. Sci. 1991, 45, 186–203. [Google Scholar]
- Randall, J.E. Zoogeography of shore fishes of the Indo-Pacific region. Zool. Stud. 1998, 37, 227–268. [Google Scholar]
- DiBattista, J.D.; Wilcox, C.; Craig, M.T.; Rocha, L.A.; Bowen, B.W. Phylogeography of the Pacific Blueline Surgeonfish Acanthurus nigroris reveals a cryptic species in the Hawaiian Archipelago. J. Mar. Biol. 2011, 2011, 839134. [Google Scholar] [CrossRef]
- Skillings, D.J.; Bird, C.E.; Toonen, R.J. Gateways to Hawai‘i: Genetic Population Structure of the Tropical Sea Cucumber Holothuria atra. J. Mar. Biol. 2010, 2011, 783030. [Google Scholar] [CrossRef]
- Wren, J.L.K.; Kobayashi, D.R.; Jia, Y.; Toonen, R.J. Modeled Population Connectivity across the Hawaiian Archipelago. PLoS ONE 2016, 11, e0167626. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, D.R.; Polovina, J.J. Simulated seasonal and interannual variability in larval transport and oceanography in the Northwestern Hawaiian Islands using satellite remotely sensed data and computer modeling. Atoll Res. Bull. 2006, 543, 365–390. [Google Scholar]
- Zeng, X.; Adams, A.; Roffer, M.; He, R. Potential connectivity among spatially distinct management zones for Bonefish (Albula vulpes) via larval dispersal. Environ. Biol. Fishes 2018, 102, 233–252. [Google Scholar] [CrossRef]
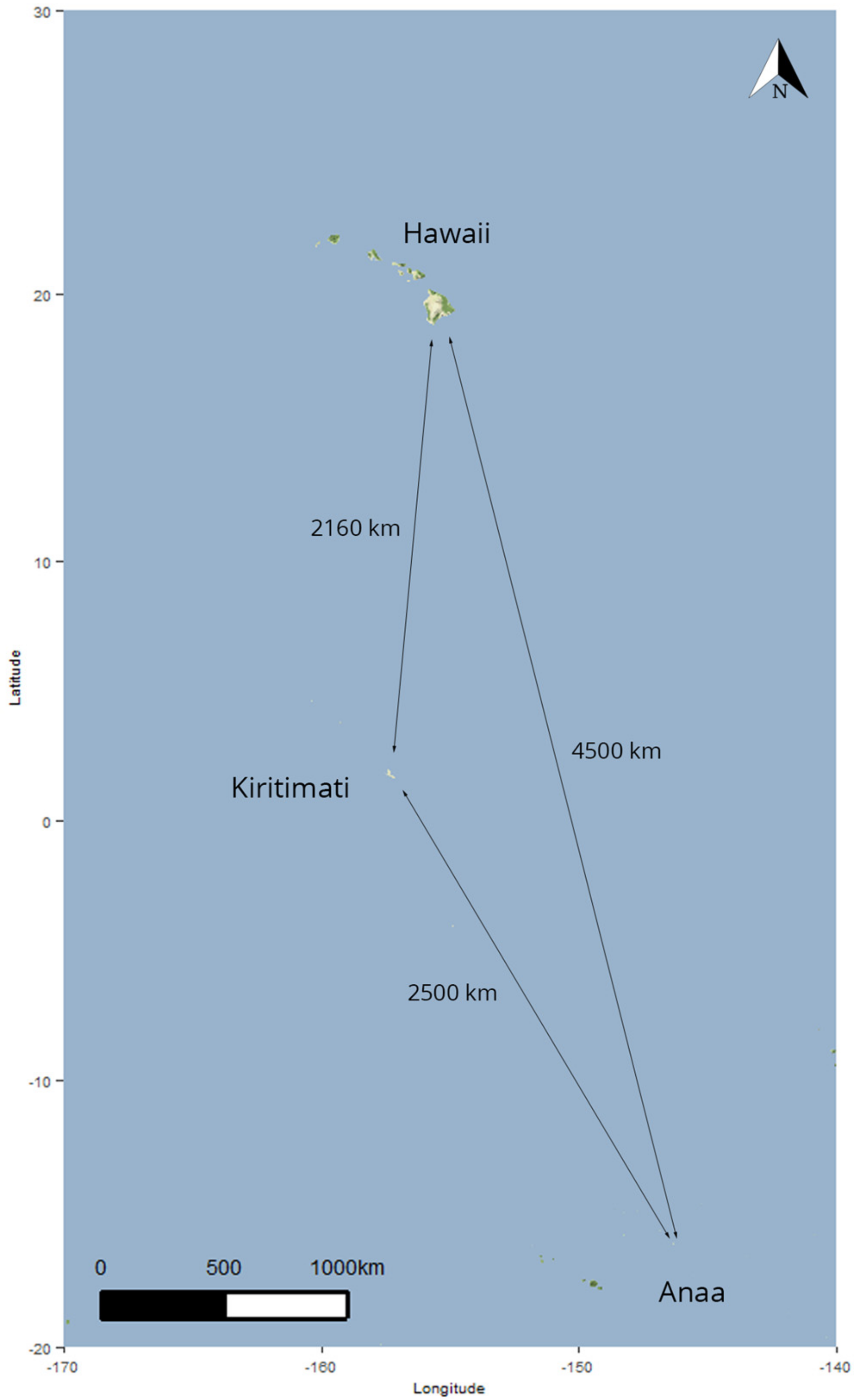
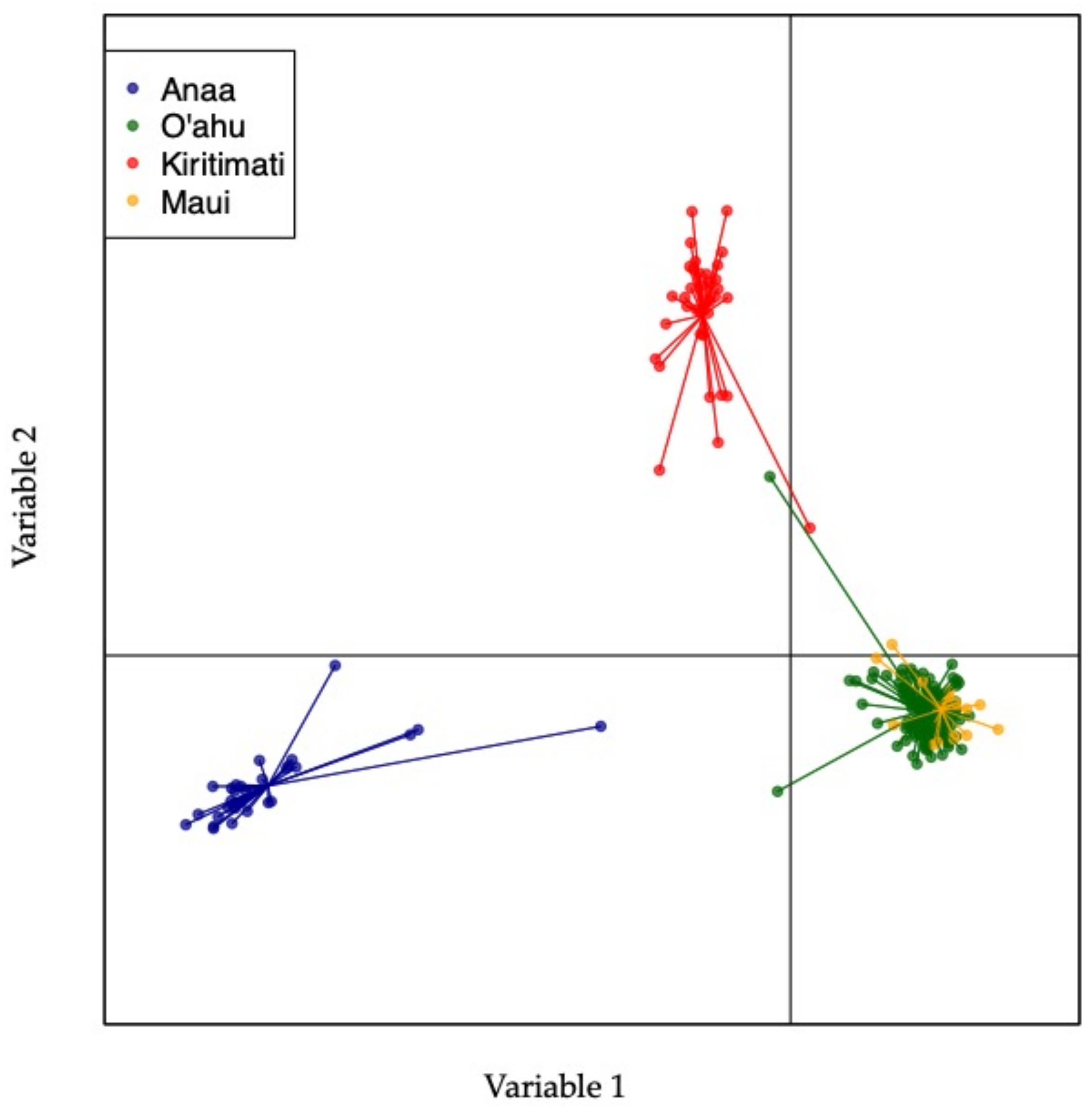
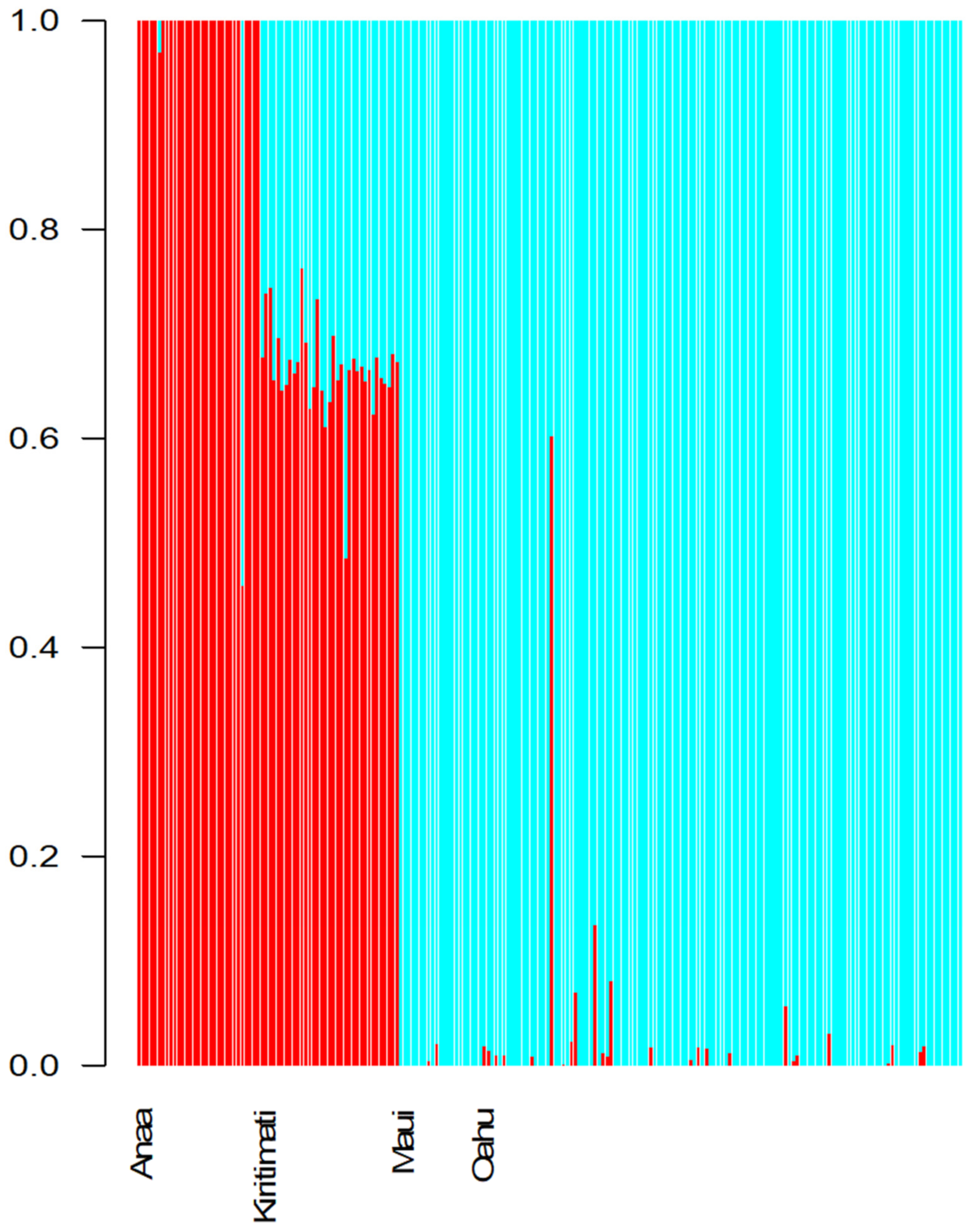

| Location | Samples Collected | Samples Processed |
|---|---|---|
| O’ahu (adults) | 150 | 108 |
| O’ahu (leptocephalus) | 28 | 23 |
| Maui | 30 | 30 |
| Kiritimati | 37 | 36 |
| Anaa | 58 | 57 |
| Total | 303 | 254 * |
| Location | N | MLG | eMLG | SE | H | G | λ | E | Hexp |
|---|---|---|---|---|---|---|---|---|---|
| Anaa | 31 | 31 | 20 | 0.00 | 3.43 | 31 | 0.97 | 1 | 0.23 |
| O’ahu | 122 | 122 | 20 | 0.00 | 4.80 | 122 | 0.99 | 1 | 0.24 |
| Kiritimati | 35 | 35 | 20 | 0.00 | 3.56 | 35 | 0.97 | 1 | 0.24 |
| Maui | 20 | 20 | 20 | 0.00 | 3.00 | 20 | 0.95 | 1 | 0.24 |
| Anaa | O’ahu | Kiritimati | Maui | |
|---|---|---|---|---|
| Anaa | ~ | 0.0998 | 0.0416 | 0.1066 |
| O’ahu | 0.0847 | ~ | 0.0509 | 0.005 |
| Kiritimati | 0.0325 | 0.0415 | ~ | 0.0537 |
| Maui | 0.0872 | 0.0016 | 0.043 | ~ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamikawa, K.; Bowen, B.W.; Kobayashi, D.; Peyton, K.; Wallace, E. Genetic Connectivity of Roundjaw Bonefish Albula glossodonta (Elopomorpha, Albulidae) in the Central Pacific Ocean Resolved through ddRAD-Based Population Genomics. Fishes 2023, 8, 585. https://doi.org/10.3390/fishes8120585
Kamikawa K, Bowen BW, Kobayashi D, Peyton K, Wallace E. Genetic Connectivity of Roundjaw Bonefish Albula glossodonta (Elopomorpha, Albulidae) in the Central Pacific Ocean Resolved through ddRAD-Based Population Genomics. Fishes. 2023; 8(12):585. https://doi.org/10.3390/fishes8120585
Chicago/Turabian StyleKamikawa, Keith, Brian W. Bowen, Donald Kobayashi, Kimberly Peyton, and Elizabeth Wallace. 2023. "Genetic Connectivity of Roundjaw Bonefish Albula glossodonta (Elopomorpha, Albulidae) in the Central Pacific Ocean Resolved through ddRAD-Based Population Genomics" Fishes 8, no. 12: 585. https://doi.org/10.3390/fishes8120585
APA StyleKamikawa, K., Bowen, B. W., Kobayashi, D., Peyton, K., & Wallace, E. (2023). Genetic Connectivity of Roundjaw Bonefish Albula glossodonta (Elopomorpha, Albulidae) in the Central Pacific Ocean Resolved through ddRAD-Based Population Genomics. Fishes, 8(12), 585. https://doi.org/10.3390/fishes8120585






