Molecular Identification and Expression Analysis of an Intelectin Gene in the Yellow Catfish Pelteobagrus fulvidraco (Siluriformes: Bagridae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction and cDNA Synthesis
2.3. Cloning PfITLN cDNA
2.4. Sequence Analysis of PfITLN
2.5. Homologous Alignment and Phylogenetic Analysis
2.6. Quantitative Analysis of PfITLN
2.7. Data Analysis
3. Results and Discussion
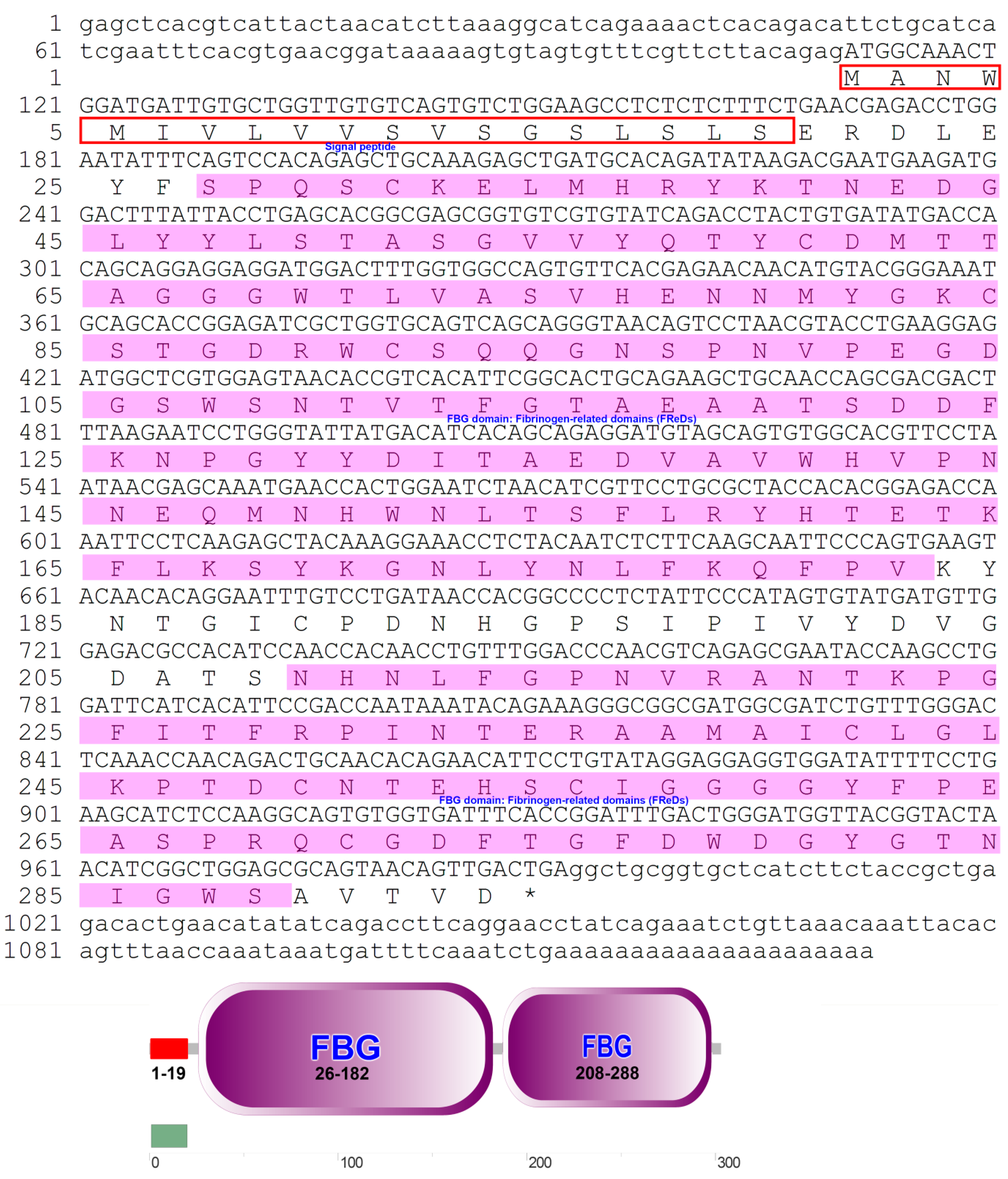
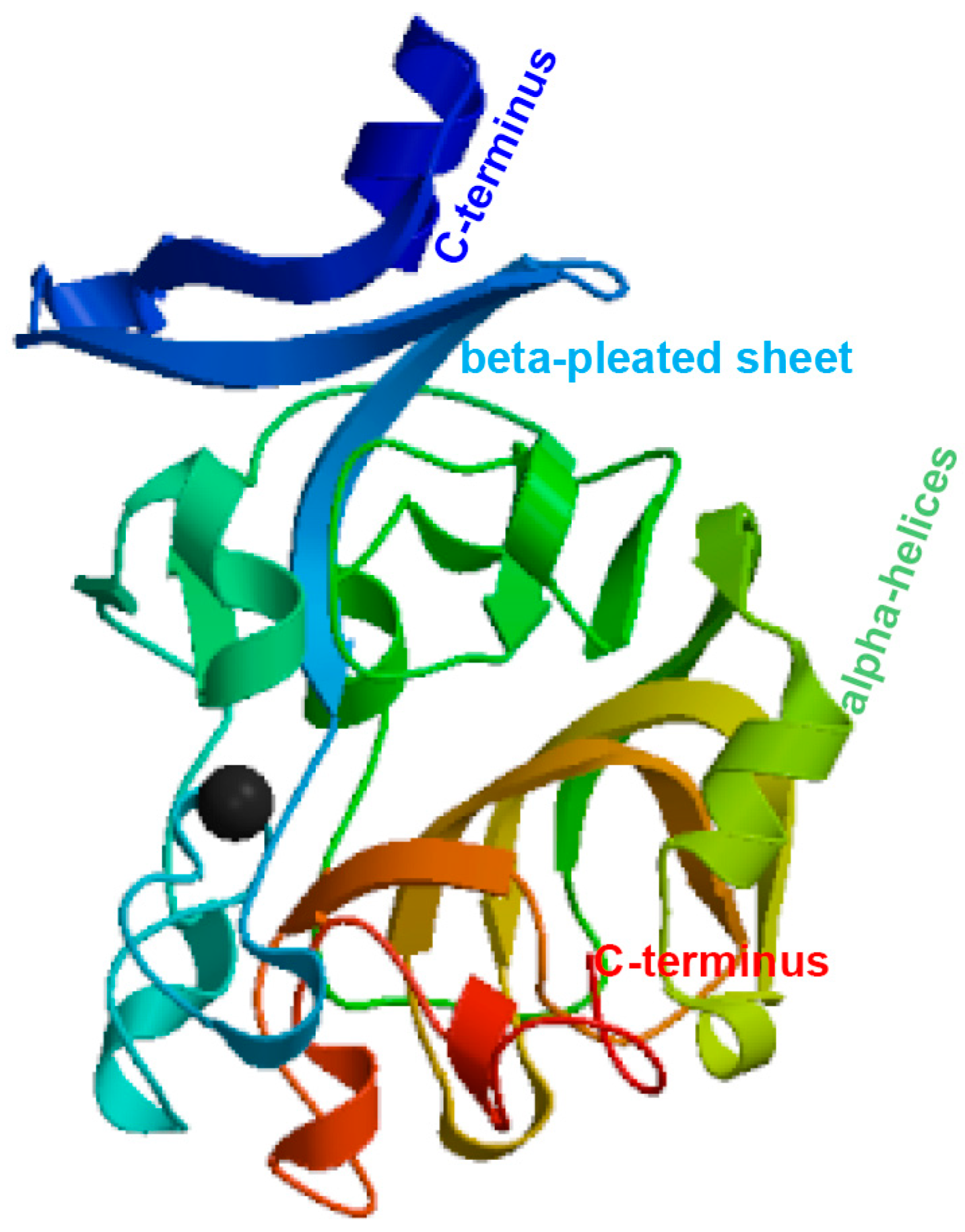
3.1. Sequence Analysis of the PfITLN Gene
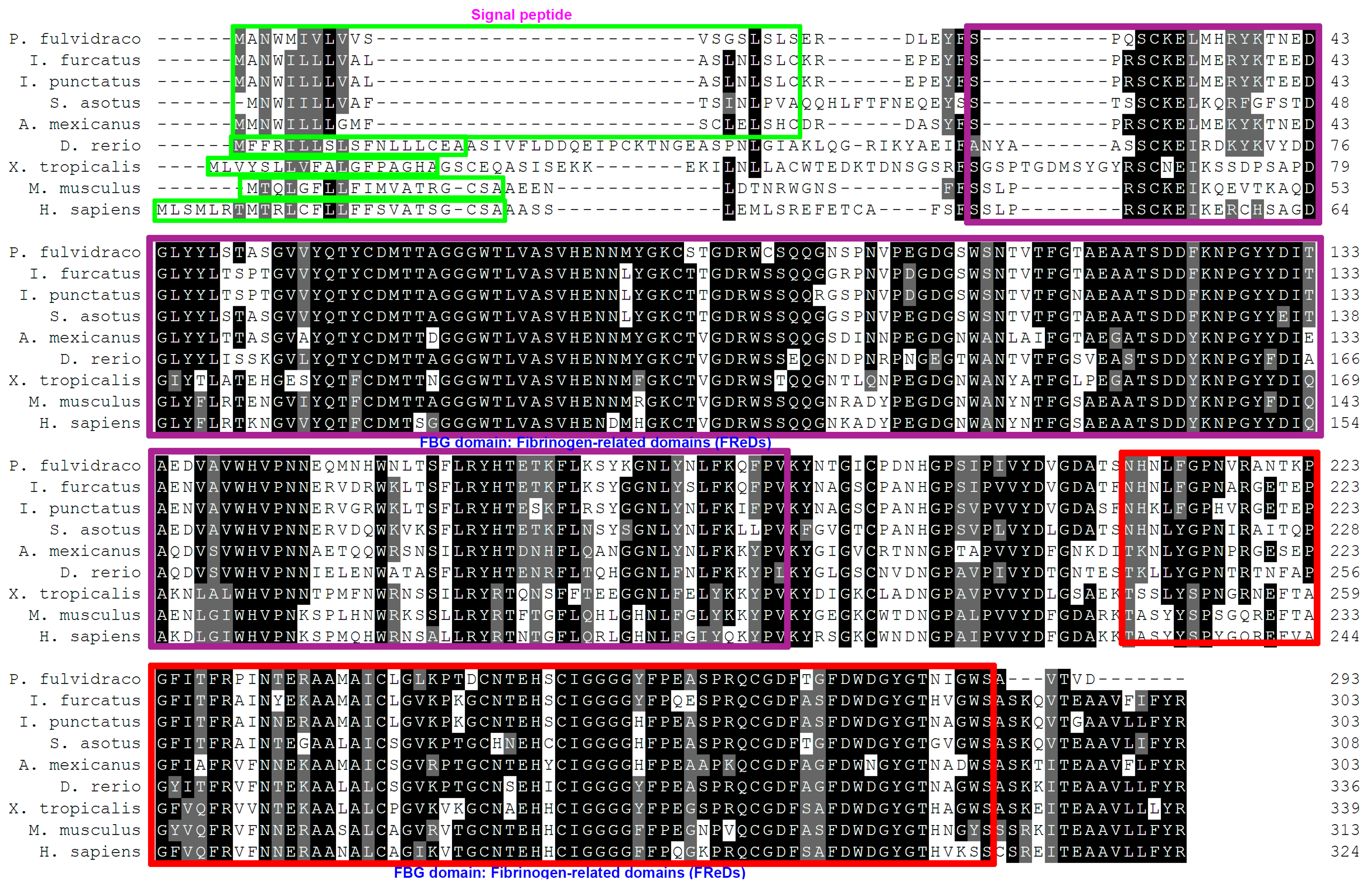
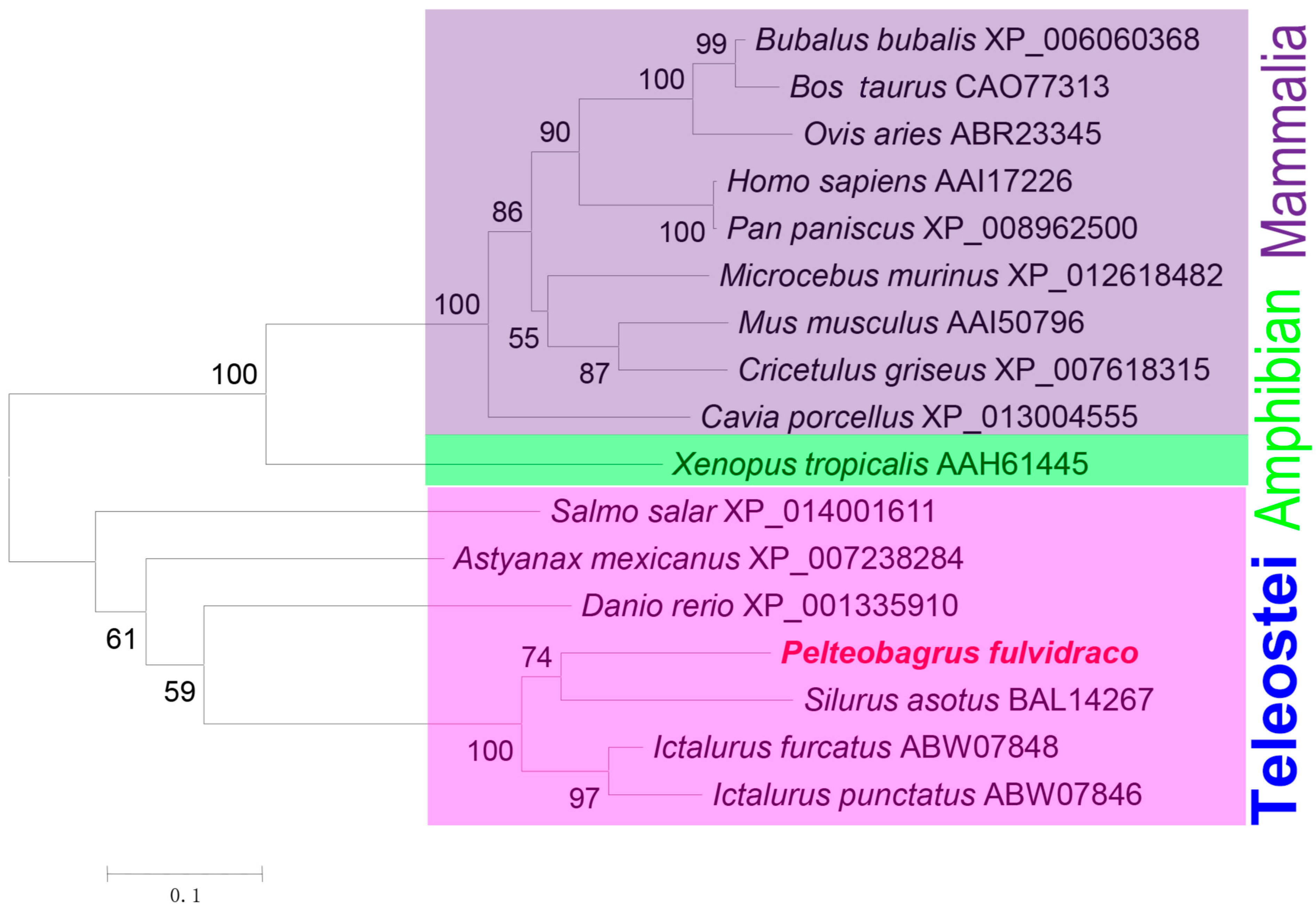
3.2. Homologous Sequence Alignment and Phylogenetic Analysis
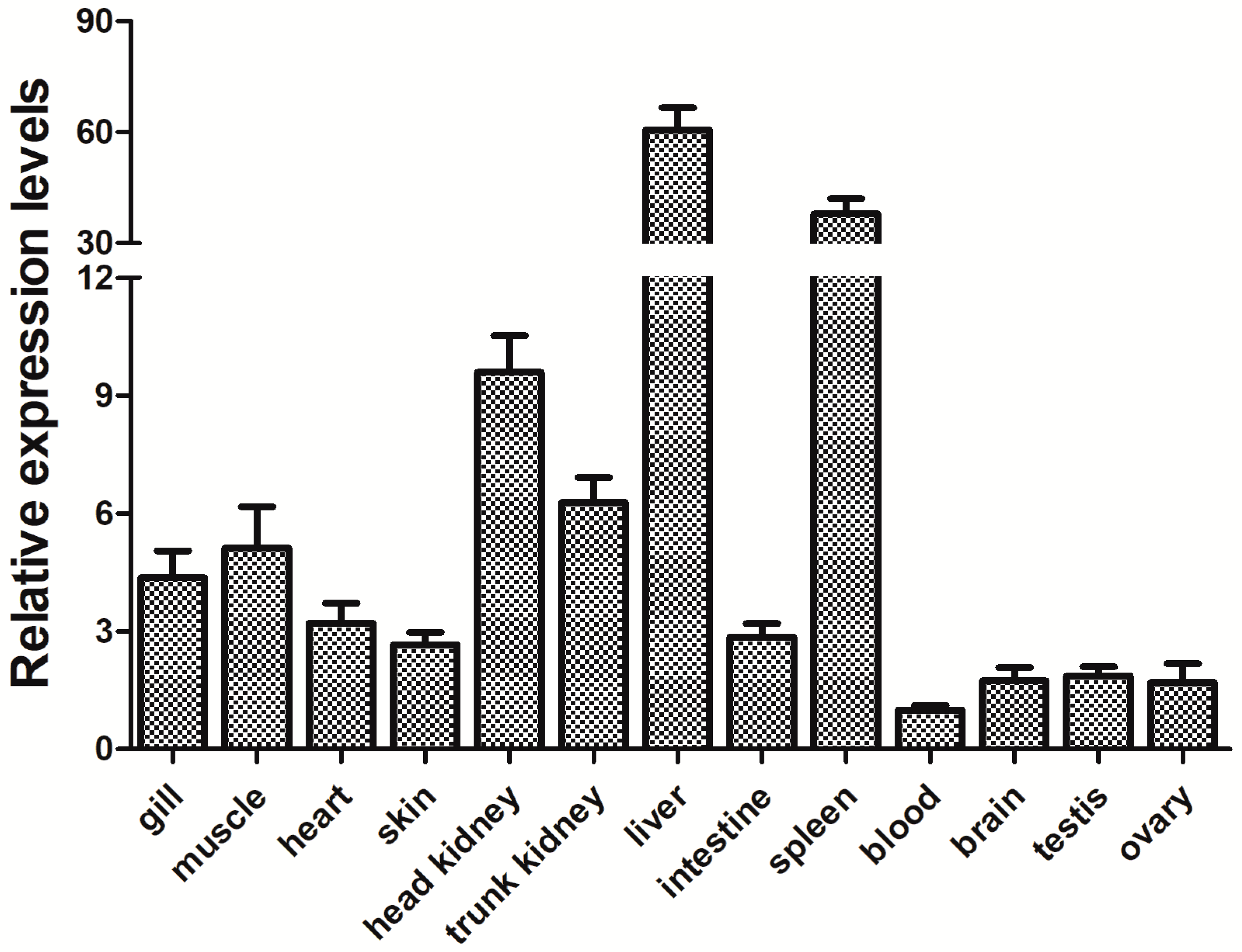
3.3. Expression of the PfITLN Gene in Yellow Catfish
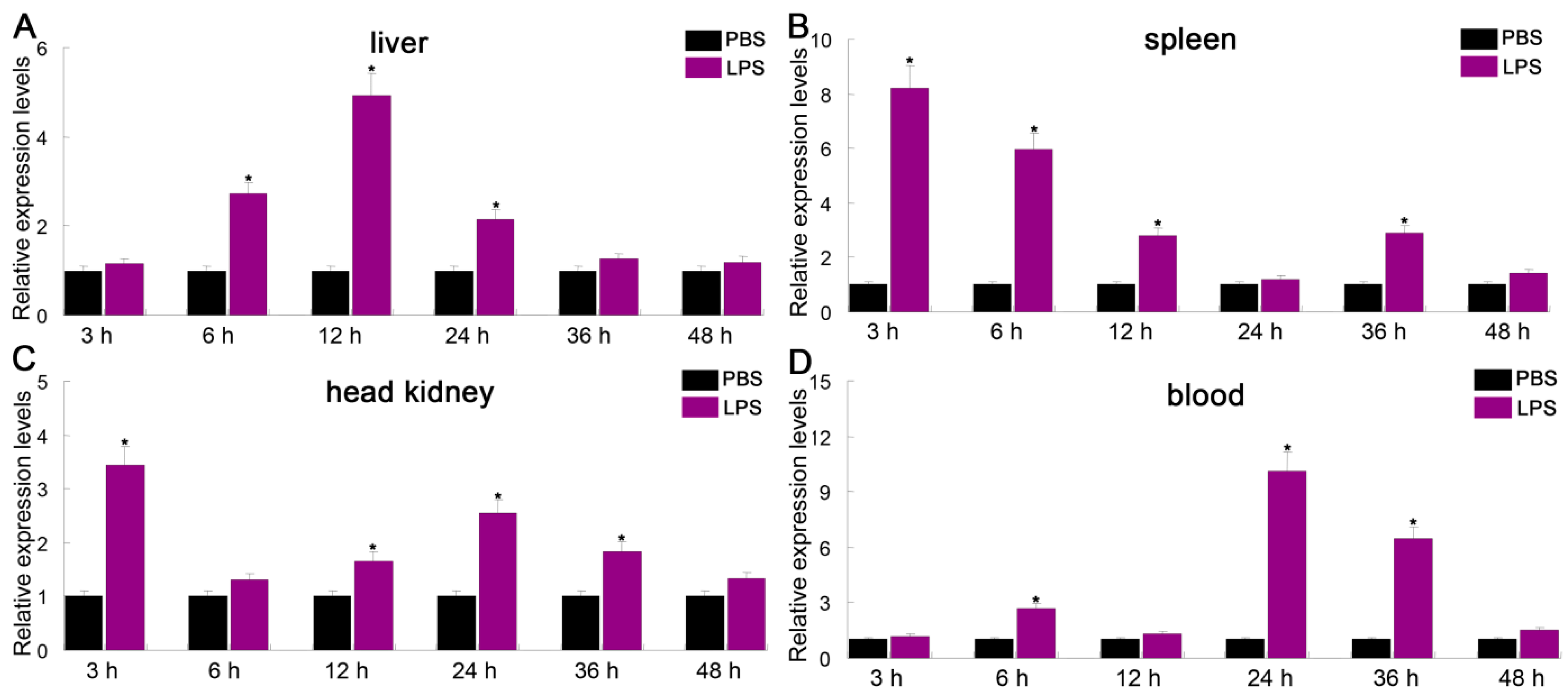
3.4. Expression Profiles of PfITLN Challenged by LPS or Poly I:C
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, L.; Nie, L.; Zhu, G.; Xiang, L.X.; Shao, J.Z. Advances in research of fish immune-relevant genes: A comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 2013, 39, 39–62. [Google Scholar] [CrossRef]
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar] [CrossRef]
- Yuan, S.; Sun, Z.; Gao, Q.; Li, Z.; Qi, Z.; Zheng, S.; Liu, D. Molecular characterization and expression analysis of NLRC3-like, ASC, and caspase1 in spotted sea bass (Lateolabrax maculatus). Fishes 2023, 8, 378. [Google Scholar] [CrossRef]
- Guan, R.; Mariuzza, R.A. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 2007, 15, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.B.; Fai Cheung, R.C.; Wing Ng, C.C.; Fang, E.F.; Wong, J.H. A review of fish lectins. Curr. Protein Pept. Sci. 2015, 16, 337–351. [Google Scholar] [CrossRef]
- Tang, X.H.; Ye, X.T.; Wu, Q.C.; Tang, Y.Y.; Zhang, D.Z.; Liu, Q.N.; Tang, B.P.; Zhu, H.R. Molecular characterization and expression analysis of a novel C-type lectin (CTL) gene in yellow catfish Pelteobagrus fulvidraco. Aquacult. Rep. 2021, 20, 100640. [Google Scholar] [CrossRef]
- Vasta, G.R.; Amzel, L.M.; Bianchet, M.A.; Cammarata, M.; Feng, C.; Saito, K. F-type lectins: A highly diversified family of fucose-binding proteins with a unique sequence motif and structural fold, involved in self/non-self-recognition. Front. Immunol. 2017, 8, 1648. [Google Scholar] [CrossRef]
- Niu, J.; Huang, Y.; Liu, X.; Wu, F.; Tang, J.; Wang, B.; Lu, Y.; Cai, J.; Jian, J. Fish galectin8-like exerts positive regulation on immune response against bacterial infection. Front. Immunol. 2020, 11, 1140. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, Y.; Li, H.; Wang, X.; Zhang, M.; Shen, X.; Cheng, H.; Xu, J.; Wang, X.; Liu, H.; et al. Novel insights into the immune regulatory effects of Megalobrama amblycephala intelectin on the phagocytosis and killing activity of macrophages. Mol. Immunol. 2021, 137, 145–154. [Google Scholar] [CrossRef]
- Sugawara, S.; Tatsuta, T.; Hosono, M. Bacterial Expression of Rhamnose-Binding Lectin from Catfish Eggs. Methods Mol. Biol. 2020, 2132, 359–367. [Google Scholar]
- Angata, T.; Brinkman-Van der Linden, E. I-type lectins. Biochim. Biophys. Acta 2002, 1572, 294–316. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, X.; Wang, Z.; Zhang, Q. Immunological characterization and function analysis of L-type lectin from spotted knifejaw, Oplegnathus punctatus. Front. Immunol. 2022, 13, 993777. [Google Scholar] [CrossRef] [PubMed]
- Wyrick, R.E.; Nishihara, T.; Hedrick, J.L. Agglutination of jelly coat and cortical granule components and the block to polyspermy in the amphibian Xenopus laevis. Proc. Natl. Acad. Sci. USA 1974, 71, 2067–2071. [Google Scholar] [CrossRef] [PubMed]
- Blanco Vázquez, C.; Balseiro, A.; Alonso-Hearn, M.; Juste, R.A.; Iglesias, N.; Canive, M.; Casais, R. Bovine intelectin 2 expression as a biomarker of paratuberculosis disease progression. Animals (Basel) 2021, 11, 1370. [Google Scholar] [CrossRef]
- French, A.T.; Bethune, J.A.; Knight, P.A.; McNeilly, T.N.; Wattegedera, S.; Rhind, S.; Miller, H.R.P.; Pemberton, A.D. The expression of intelectin in sheep goblet cells and upregulation by interleukin-4. Vet. Immunol. Immunopathol. 2007, 120, 41–46. [Google Scholar] [CrossRef]
- Kuperman, D.; Lewis, C.; Woodruff, P.; Rodriguez, M.W.; Yang, Y.H.; Dolganov, G.M.; Fahy, J.V.; Erle, D.J. Dissecting asthma using focused transgenic modelling and functional genomics. J. Allergy Clin. Immunol. 2005, 116, 305–311. [Google Scholar] [CrossRef]
- Akiyama, Y.; Oshima, K.; Kuhara, T.; Shin, K.; Abe, F.; Iwatsuki, K.; Nadano, D.; Matsuda, T. A lactoferrin-receptor, intelectin 1, affects uptake, sub-cellular localization and release of immunochemically detectable lactoferrin by intestinal epithelial Caco-2 cells. J. Biochem. 2013, 154, 437–448. [Google Scholar] [CrossRef]
- Washimi, K.; Yokose, T.; Yamashita, M.; Kageyama, T.; Suzuki, K.; Yoshihara, M.; Miyagi, Y.; Hayashi, H.; Tsuji, S. Specific expression of human intelectin-1 in malignant pleural mesothelioma and gastrointestinal goblet cells. PLoS ONE 2012, 7, e39889. [Google Scholar] [CrossRef]
- Lee, J.K.; Buckhaults, P.; Wilkes, C.; Teilhet, M.; King, M.L.; Moremen, K.W.; Pierce, M. Cloning and expression of a Xenopus laevis oocyte lectin and characterization of its mRNA levels during early development. Glycobiology 1997, 7, 367–372. [Google Scholar] [CrossRef]
- Tsuji, S.; Uehori, J.; Matsumoto, M.; Suzuki, Y.; Matsuhisa, A.; Toyoshima, K.; Seya, T. Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell walls. J. Biol. Chem. 2001, 276, 23456–23463. [Google Scholar] [CrossRef]
- Nagata, S. Xenopus laevis macrophage-like cells produce XCL-1, an intelectin family serum lectin that recognizes bacteria. Immunol. Cell Biol. 2018, 96, 872–878. [Google Scholar] [CrossRef]
- Cui, H.; Liu, Y.; Zheng, Y.; Li, H.; Zhang, M.; Wang, X.; Zhao, X.; Cheng, H.; Xu, J.; Chen, X.; et al. Intelectin enhances the phagocytosis of macrophages via CDC42-WASF2-ARPC2 signalling axis in Megalobrama amblycephala. Int. J. Biol. Macromol. 2023, 236, 124027. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Yang, M.; Jiang, H.; Li, T.; Duan, G.; Ling, J.; Gao, Q. Effect of astragalus membranaceus on transcriptome and survival of hybrid yellow catfish (Pseudobagrus vachellii ♂ × Tachysurus fulvidraco ♀) in response to Aeromonas hydrophila challenge. Fishes 2023, 8, 454. [Google Scholar] [CrossRef]
- Wesener, D.A.; Dugan, A.; Kiessling, L.L. Recognition of microbial glycans by soluble human lectins. Curr. Opin. Struct. Biol. 2017, 44, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, J.; Yang, G. A comparative review of interactions. Scand J. Immunol. 2020, 92, e12882. [Google Scholar] [CrossRef]
- Ding, Z.; Zhao, X.; Wang, J.; Zhang, F.; Wang, W.; Liu, H. Intelectin mediated phagocytosis and killing activity of macrophages in blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol. 2019, 87, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Singrang, N.; Sitthiyotha, T.; Chomanee, N.; Watthanasak, C.; Chunsrivirot, S.; Wangkanont, K. Molecular properties and ligand specificity of zebrafish intelectin-2. Fish Shellfish Immunol. 2022, 123, 528–536. [Google Scholar] [CrossRef]
- Russell, S.; Hayes, M.A.; Lumsden, J.S. Immunohistochemical localization of rainbow trout ladder lectin and intelectin in healthy and infected rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2009, 26, 154–163. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Gu, W.; Xu, F.; Li, H.; Shan, S.; Sun, X.; Yin, M.; Yang, G.; Chen, L. Characterization of a common carp intelectin gene with bacterial binding and agglutination activity. Fish Shellfish Immunol. 2021, 108, 32–41. [Google Scholar] [CrossRef]
- Chang, M.X.; Nie, P. Intelectin gene from the grass carp Ctenopharyngodon idella: cDNA cloning, tissue expression, and immunohistochemical localization. Fish Shellfish Immunol. 2007, 23, 128–140. [Google Scholar] [CrossRef]
- Meiers, J.; Siebs, E.; Zahorska, E.; Titz, A. Lectin antagonists in infection, immunity, and inflammation. Curr. Opin. Chem. Biol. 2019, 53, 51–67. [Google Scholar] [CrossRef]
- Liu, H.; Guan, B.; Xu, J.; Hou, C.; Tian, H.; Chen, H. Genetic manipulation of sex ratio for the large-scale breeding of YY super-male and XY all-male yellow catfish (Pelteobagrus fulvidraco (Richardson)). Mar. Biotechnol. 2013, 15, 321–328. [Google Scholar] [CrossRef]
- Gao, D.; Huang, J.; Lin, G.; Lu, J. Time-course transcriptome analysis of gonads from yellow catfish (Pelteobagrus fulvidraco) reveals genes associated with gonad development. BMC Genom. 2022, 23, 409. [Google Scholar] [CrossRef]
- Liu, G.H.; Zhang, D.G.; Lei, X.J.; Tan, X.Y.; Song, C.C.; Zheng, H.; Luo, Z. Effects of dietary selenium and oxidized fish oils on intestinal lipid metabolism and antioxidant responses of yellow catfish Pelteobagrus fulvidraco. Antioxidants 2022, 11, 1904. [Google Scholar] [CrossRef]
- Gong, G.; Dan, C.; Xiao, S.; Guo, W.; Huang, P.; Xiong, Y.; Wu, J.; He, Y.; Zhang, J.; Li, X.; et al. Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis. Gigascience 2018, 7, giy120. [Google Scholar] [CrossRef]
- Chen, Q.L.; Gong, Y.; Luo, Z.; Zheng, J.L.; Zhu, Q.L. Differential effect of waterborne cadmium exposure on lipid metabolism in liver and muscle of yellow catfish Pelteobagrus fulvidraco. Aquat. Toxicol. 2013, 142–143, 380–386. [Google Scholar] [CrossRef]
- Liu, Y.; Xin, Z.Z.; Zhang, D.Z.; Wang, Z.F.; Zhu, X.Y.; Tang, B.P.; Jiang, S.-H.; Zhang, H.-B.; Zhou, C.-L.; Chai, X.-Y.; et al. Transcriptome analysis of yellow catfish (Pelteobagrus fulvidraco) liver challenged with polyriboinosinic polyribocytidylic acid (poly I:C). Fish Shellfish Immunol. 2017, 68, 395–403. [Google Scholar] [CrossRef]
- Liu, Q.N.; Xin, Z.Z.; Liu, Y.; Zhang, D.Z.; Jiang, S.H.; Chai, X.Y.; Wang, Z.F.; Zhang, H.-B.; Bian, X.-G.; Zhou, C.-L.; et al. De novo transcriptome assembly and analysis of differential gene expression following lipopolysaccharide challenge in Pelteobagrus fulvidraco. Fish Shellfish Immunol. 2018, 73, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.H.; Wu, L.X.; Cai, Y.T.; Ma, R.T.; Zhang, H.B.; Zhang, D.Z.; Tang, B.P.; Liu, Q.N.; Dai, L.S. Differentially expressed genes in head kidney of Pelteobagrus fulvidraco following Vibrio cholerae challenge. Front. Immunol. 2023, 13, 1039956. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.N.; Tang, Y.Y.; Zhou, M.J.; Luo, S.; Li, Y.T.; Wang, G.; Zhang, D.Z.; Yang, H.; Tang, B.P.; He, W.F. Differentially expressed genes involved in immune pathways from yellowhead catfish (Tachysurus fulvidraco) after poly (I:C) challenge. Int. J. Biol. Macromol. 2021, 183, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wesener, D.A.; Wangkanont, K.; McBride, R.; Song, X.; Kraft, M.B.; Hodges, H.L.; Zarling, L.C.; Splain, R.A.; Smith, D.F.; Cummings, R.D.; et al. Recognition of microbial glycans by human intelectin-1. Nat. Struct. Mol. Biol. 2015, 22, 603–610. [Google Scholar] [CrossRef]
- Yan, J.; Xu, L.; Zhang, Y.; Zhang, C.; Zhang, C.; Zhao, F.; Feng, L. Comparative genomic and phylogenetic analyses of the intelectin gene family: Implications for their origin and evolution. Dev. Comp. Immunol. 2013, 41, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, K.; Maeda, K.; Saigo, C.; Kito, Y.; Yoshida, K.; Takeuchi, T. Adiponectin and intelectin-1: Important adipokine players in obesity-related colorectal carcinogenesis. Int. J. Mol. Sci. 2017, 18, 866. [Google Scholar] [CrossRef]
- Ding, Z.; Zhao, X.; Zhan, Q.; Cui, L.; Sun, Q.; Lin, L.; Wang, W.; Liu, H. Characterization and expression analysis of an intelectin gene from Megalobrama amblycephala with excellent bacterial binding and agglutination activity. Fish Shellfish Immunol. 2017, 61, 100–110. [Google Scholar] [CrossRef]
- Chen, L.; Yan, J.; Sun, W.; Zhang, Y.; Sui, C.; Qi, J.; Du, Y.; Feng, L. A zebrafish intelectin ortholog agglutinates Gram-negative and Gram-positive bacteria with binding capacity to bacterial polysaccharideS. Fish Shellfish Immunol. 2016, 55, 729–736. [Google Scholar] [CrossRef]
- Takano, T.; Sha, Z.; Peatman, E.; Terhune, J.; Liu, H.; Kucuktas, H.; Li, P.; Edholm, E.-S.; Wilson, M.; Liu, Z. The two channel catfish intelectin genes exhibit highly differential patterns of tissue expression and regulation after infection with Edwardsiella ictaluri. Dev. Comp. Immunol. 2008, 32, 693–705. [Google Scholar] [CrossRef]
- Qiao, G.; Zhang, M.; Li, Y.; Xu, C.; Xu, D.H.; Zhao, Z.; Zhang, J.; Li, Q. Biofloc technology (BFT): An alternative aquaculture system for prevention of Cyprinid herpesvirus 2 infection in gibel carp (Carassius auratus gibelio). Fish Shellfish Immunol. 2018, 83, 140–147. [Google Scholar] [CrossRef]
- Xue, Z.; Pang, Y.; Liu, X.; Zheng, Z.; Xiao, R.; Jin, M.; Han, Y.; Su, P.; Lv, L.; Wang, J.; et al. First evidence of protein G-binding protein in the most primitive vertebrate: Serum lectin from lamprey (Lampetra japonica). Dev. Comp. Immunol. 2013, 41, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, C.; Zhang, Y.; Li, K.; Xu, L.; Guo, L.; Kong, Y.; Feng, L. Characterization and comparative analyses of two amphioxus intelectins involved in the innate immune response. Fish Shellfish Immunol. 2013, 34, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Nonnecke, E.B.; Castillo, P.A.; Dugan, A.E.; Almalki, F.; Underwood, M.A.; De La Motte, C.A.; Yuan, W.; Lu, W.; Shen, B.; Johansson, M.E.V.; et al. Human intelectin-1 (ITLN1) genetic variation and intestinal expression. Sci. Rep. 2021, 11, 12889. [Google Scholar] [CrossRef]
- Nonnecke, E.B.; Castillo, P.A.; Johansson, M.E.V.; Hollox, E.J.; Shen, B.; Lönnerdal, B.; Bevins, C.L. Human intelectin-2 (ITLN2) is selectively expressed by secretory Paneth cells. FASEB J. 2022, 36, e22200. [Google Scholar] [CrossRef]
- Lu, Z.H.; di Domenico, A.; Wright, S.H.; Knight, P.A.; Whitelaw, C.B.; Pemberton, A.D. Strain-specific copy number variation in the intelectin locus on the 129 mouse chromosome 1. BMC Genom. 2011, 12, 110. [Google Scholar] [CrossRef] [PubMed]
- Fänge, R.; Nilsson, S. The fish spleen: Structure and function. Experientia 1985, 41, 152–158. [Google Scholar] [CrossRef]
- Yan, J.; Wang, J.; Zhao, Y.; Zhang, J.; Bai, C.; Zhang, C.; Zhang, C.; Li, K.; Zhang, H.; Du, X.; et al. Identification of an amphioxus intelectin homolog that preferably agglutinates gram-positive over gram-negative bacteria likely due to different binding capacity to LPS and PGN. Fish Shellfish Immunol. 2012, 33, 11–20. [Google Scholar] [CrossRef]
- Gerwick, L.; Corley-Smith, G.; Bayne, C.J. Gene transcript changes in individual rainbow trout livers following an inflammatory stimulus. Fish Shellfish Immunol. 2007, 22, 157–171. [Google Scholar] [CrossRef]
- Pemberton, A.D.; Knight, P.A.; Gamble, J.; Colledge, W.H.; Lee, J.K.; Pierce, M.; Miller, H.R. Innate BALB/c enteric epithelial responses to Trichinella spiralis: Inducible expression of a novel goblet cell lectin, intelectin-2, and its natural deletion in C57BL/10 mice. J. Immunol. 2004, 173, 1894–1901. [Google Scholar] [CrossRef]
- Bai, L.; Yu, G.; Liu, Y.; Aizaz, M.; Yang, G.; Chen, L. Common carp intelectin 3 (cITLN3) plays a role in the innate immune response. Fish Shellfish Immunol. 2023, 141, 109057. [Google Scholar] [CrossRef]
- Andresen, S.; Fantone, K.; Chapla, D.; Rada, B.; Moremen, K.W.; Pierce, M.; Szymanski, C.M. Human intelectin-1 promotes cellular attachment and neutrophil killing of Streptococcus pneumoniae in a serotype-dependent manner. Infect Immun. 2022, 90, e0068221. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, S.; Yamashita, M.; Hoffman, D.R.; Nishiyama, A.; Shinohara, T.; Ohtsu, T.; Shibata, Y. Capture of heat-killed Mycobacterium bovis bacillus Calmette-Guérin by intelectin-1 deposited on cell surfaces. Glycobiology 2009, 19, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, A.D.; Verdon, B.; Inglis, N.F.; Pearson, J.P. Sheep intelectin-2 co-purifies with the mucin Muc5ac from gastric mucus. Res. Vet. Sci. 2011, 91, e53–e57. [Google Scholar] [CrossRef]
- Nagata, S. Identification and characterization of a novel intelectin in the digestive tract of Xenopus laevis. Dev. Comp. Immunol. 2016, 59, 229–239. [Google Scholar] [CrossRef] [PubMed]

| Primer No | Primer Sequences (5′-3′) | Purpose |
|---|---|---|
| RC5 | TCTTATATCTGTGCATCAGCT | RACE-PCR |
| RC3 | TCTCTTCAAGCAATTCCCAGT | RACE-PCR |
| F1 | TGAAGGAGATGGCTCGTGGAG | qRT-PCR |
| R1 | GGGCCGTGGTTATCAGGACA | qRT-PCR |
| Actin-F | GCACAGTAAAGGCGTTGTGA | qRT-PCR |
| Actin-R | ACATCTGCTGGAAGGTGGAC | qRT-PCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Lei, Y.; Li, Y.; Sun, W.; Wang, T.; Ma, R.; Liu, Q.; Tang, B. Molecular Identification and Expression Analysis of an Intelectin Gene in the Yellow Catfish Pelteobagrus fulvidraco (Siluriformes: Bagridae). Fishes 2023, 8, 492. https://doi.org/10.3390/fishes8100492
Jiang S, Lei Y, Li Y, Sun W, Wang T, Ma R, Liu Q, Tang B. Molecular Identification and Expression Analysis of an Intelectin Gene in the Yellow Catfish Pelteobagrus fulvidraco (Siluriformes: Bagridae). Fishes. 2023; 8(10):492. https://doi.org/10.3390/fishes8100492
Chicago/Turabian StyleJiang, Senhao, Yuting Lei, Yanxuan Li, Wanyan Sun, Ti Wang, Ruiting Ma, Qiuning Liu, and Boping Tang. 2023. "Molecular Identification and Expression Analysis of an Intelectin Gene in the Yellow Catfish Pelteobagrus fulvidraco (Siluriformes: Bagridae)" Fishes 8, no. 10: 492. https://doi.org/10.3390/fishes8100492
APA StyleJiang, S., Lei, Y., Li, Y., Sun, W., Wang, T., Ma, R., Liu, Q., & Tang, B. (2023). Molecular Identification and Expression Analysis of an Intelectin Gene in the Yellow Catfish Pelteobagrus fulvidraco (Siluriformes: Bagridae). Fishes, 8(10), 492. https://doi.org/10.3390/fishes8100492






