Comparative Transcriptomics Reveals the microRNA-Mediated Immune Response of Large Yellow Croaker (Larimichthys crocea) to Pseudomonas plecoglossicida Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. L. crocea Challenge and Sampling
2.2. RNA Extraction, Library Construction, and Comparative Transcriptome Sequencing
2.3. Identification of DEmRNAs and DEmiRNAs
2.4. Association Analysis and Construction of the miRNA-mRNA Regulatory Network
2.5. Verification of Novel miRNAs, DEmiRNAs, and DEmRNAs
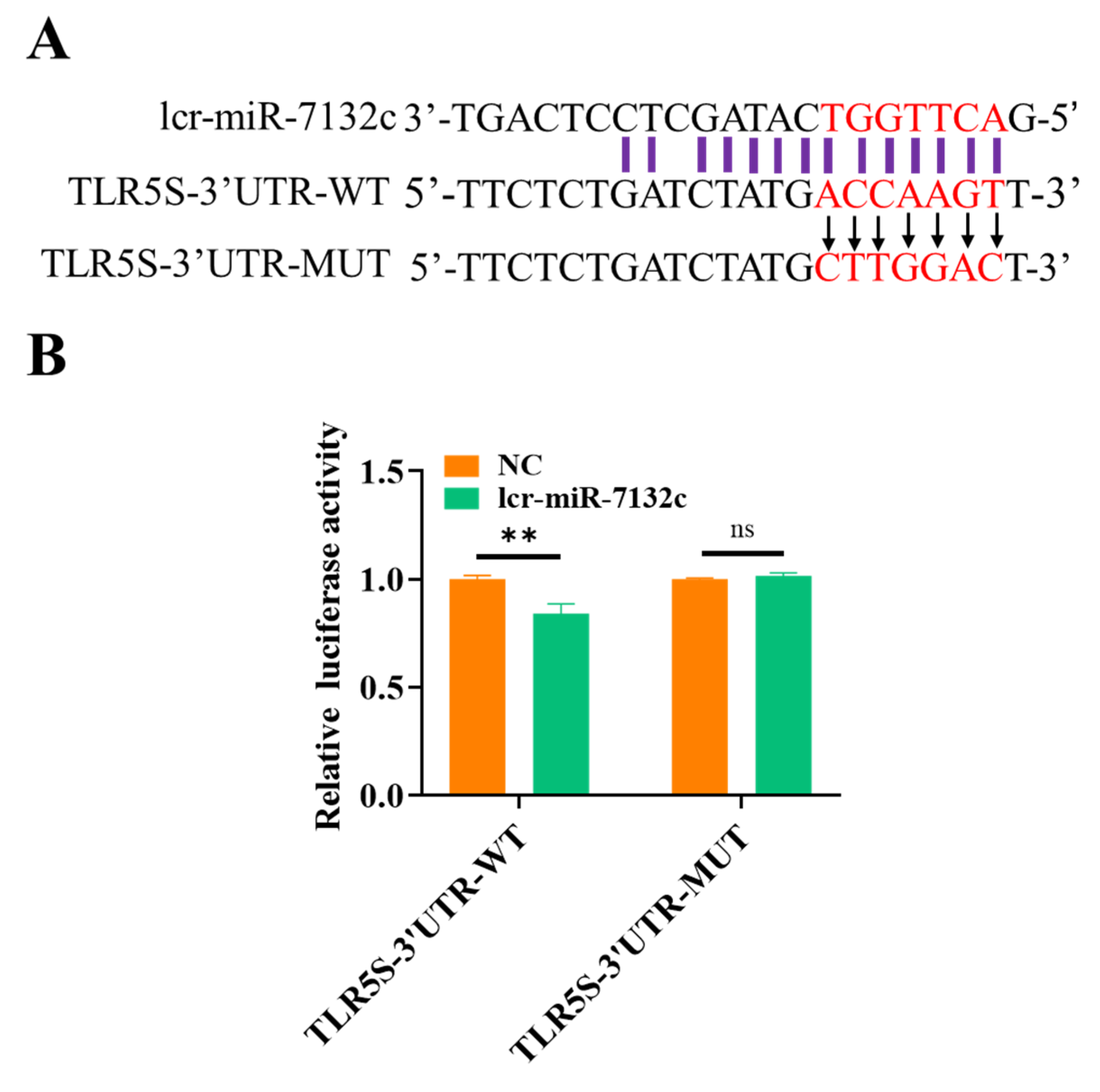
2.6. Dual-Luciferase Reporter Analysis
2.7. Statistical Analysis
3. Results
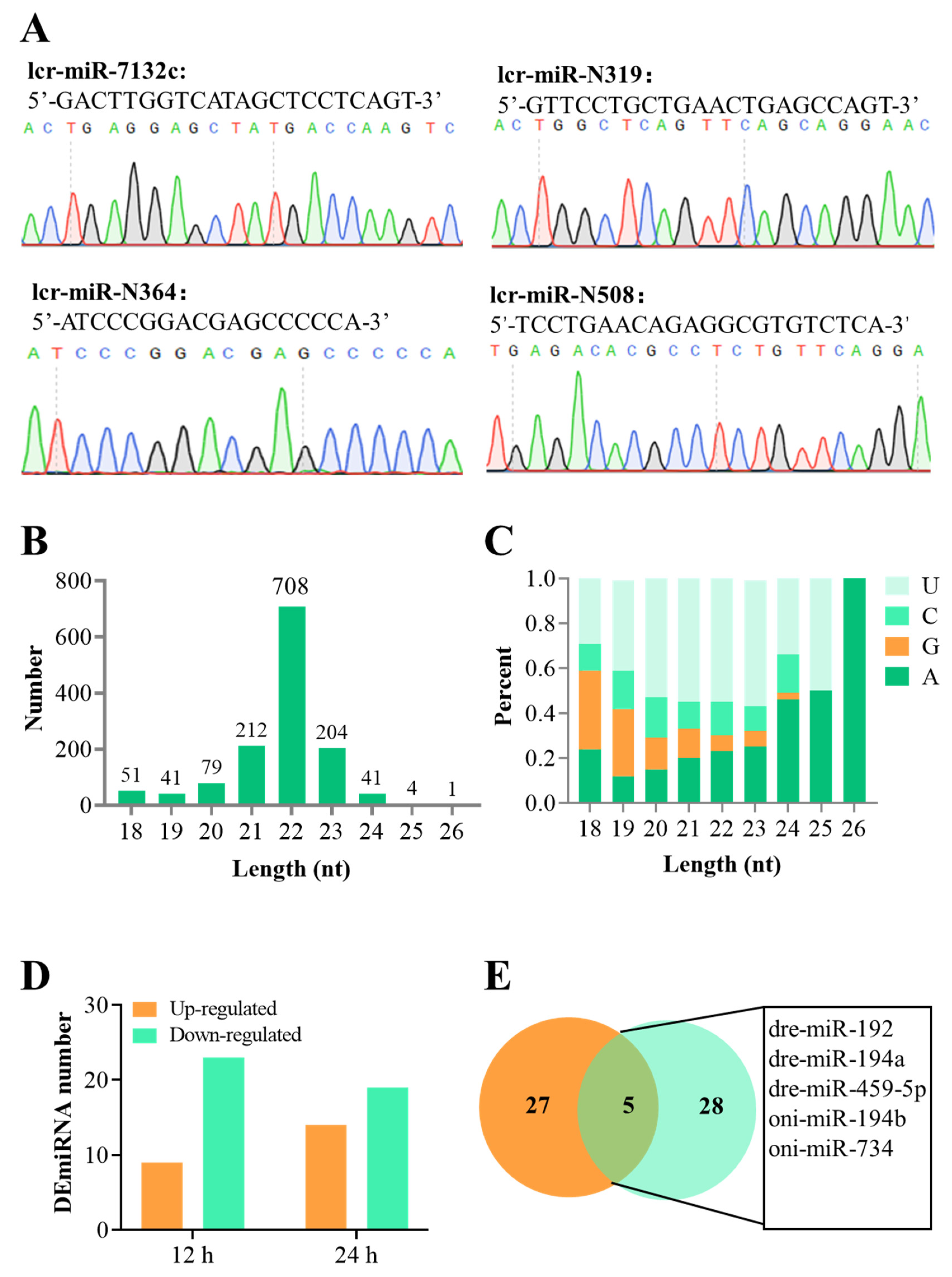
3.1. Overview of the sRNA-Seq Data and the DEmiRNAs
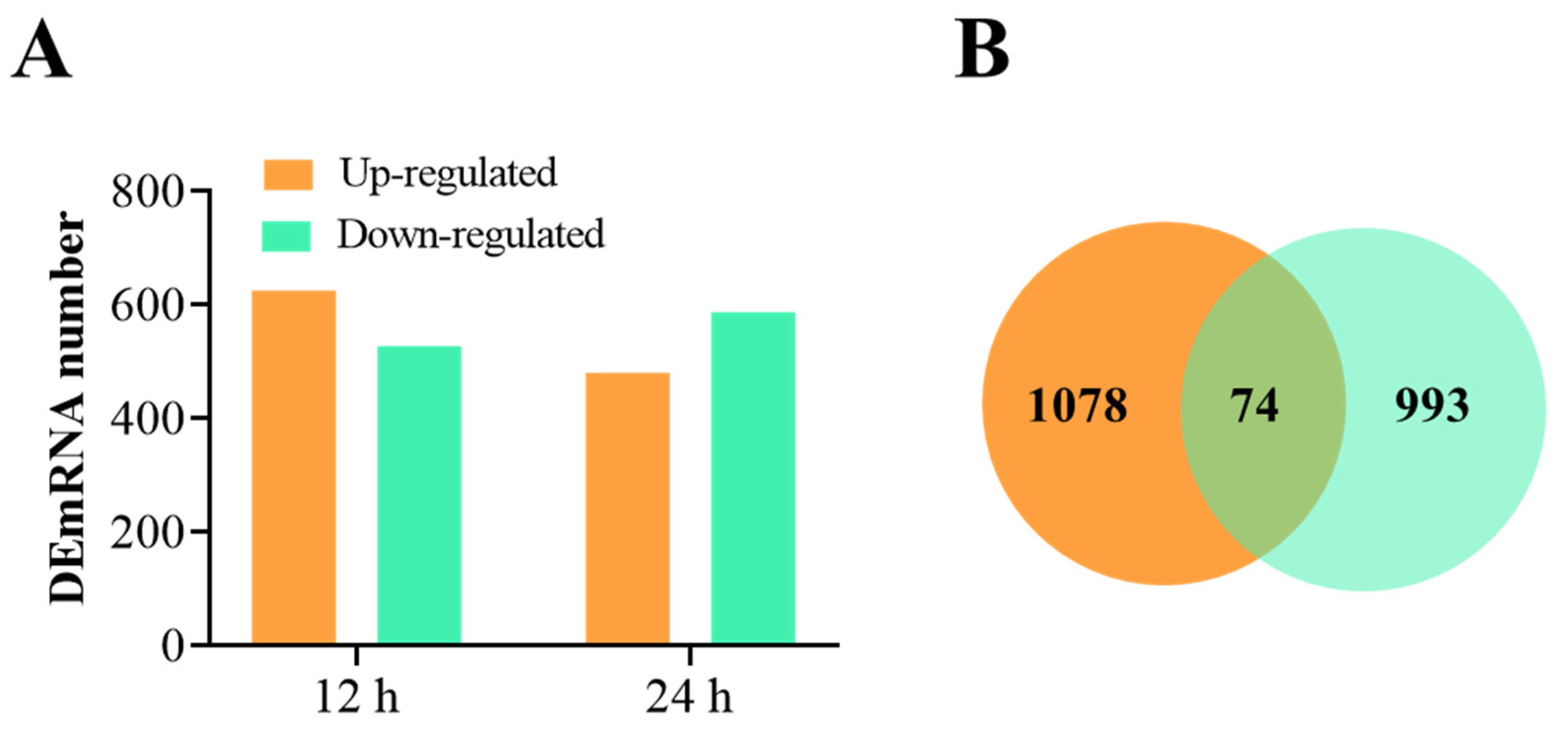
3.2. Overview of RNA-Seq Data and the DEmRNAs
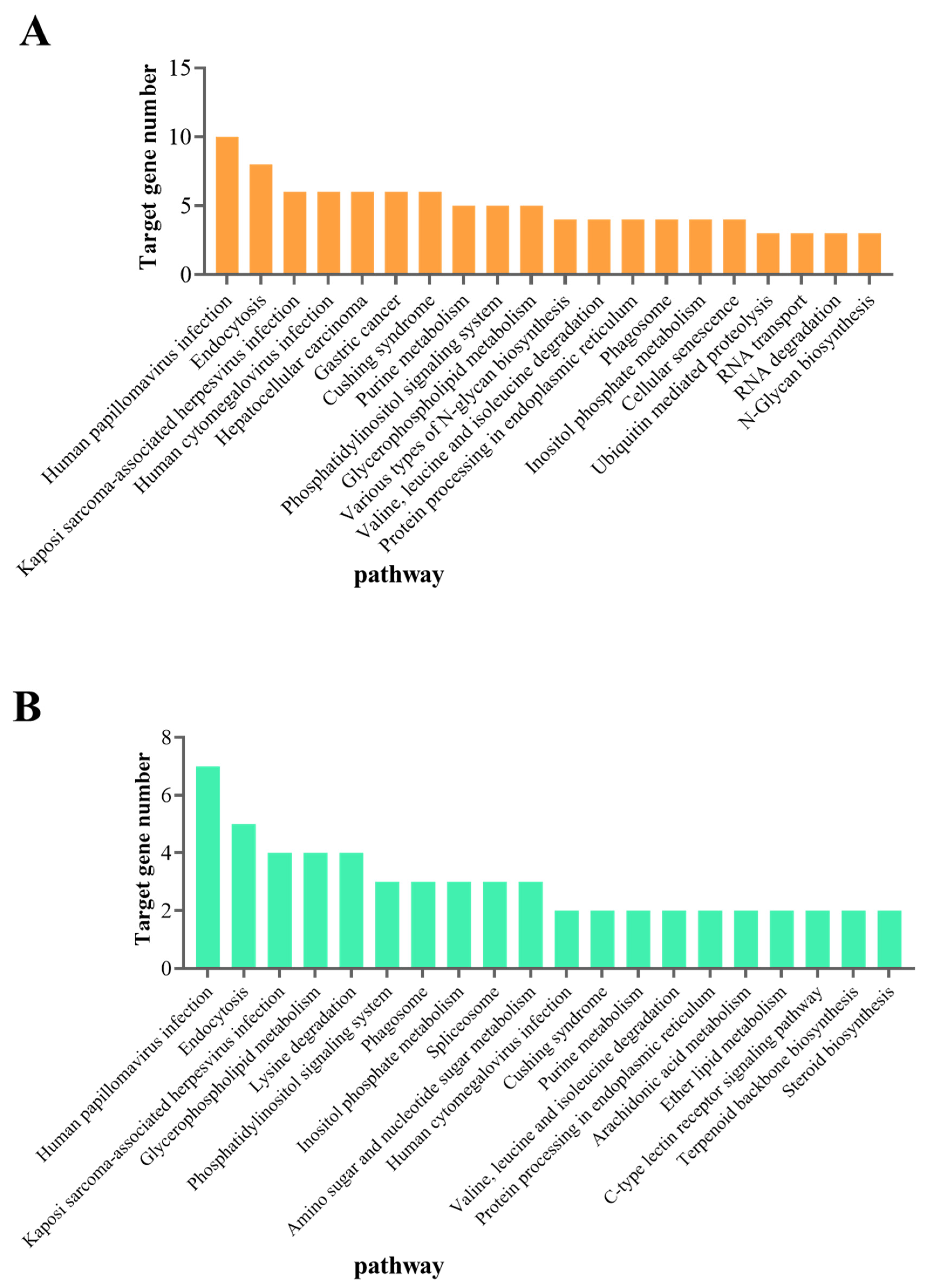
3.3. Prediction and Functional Analysis of DEmiRNA Target Genes
3.4. Functional Analysis of the DEmRNAs
3.5. Integrated Analysis of the DEmiRNA-DEmRNA Regulatory Networks
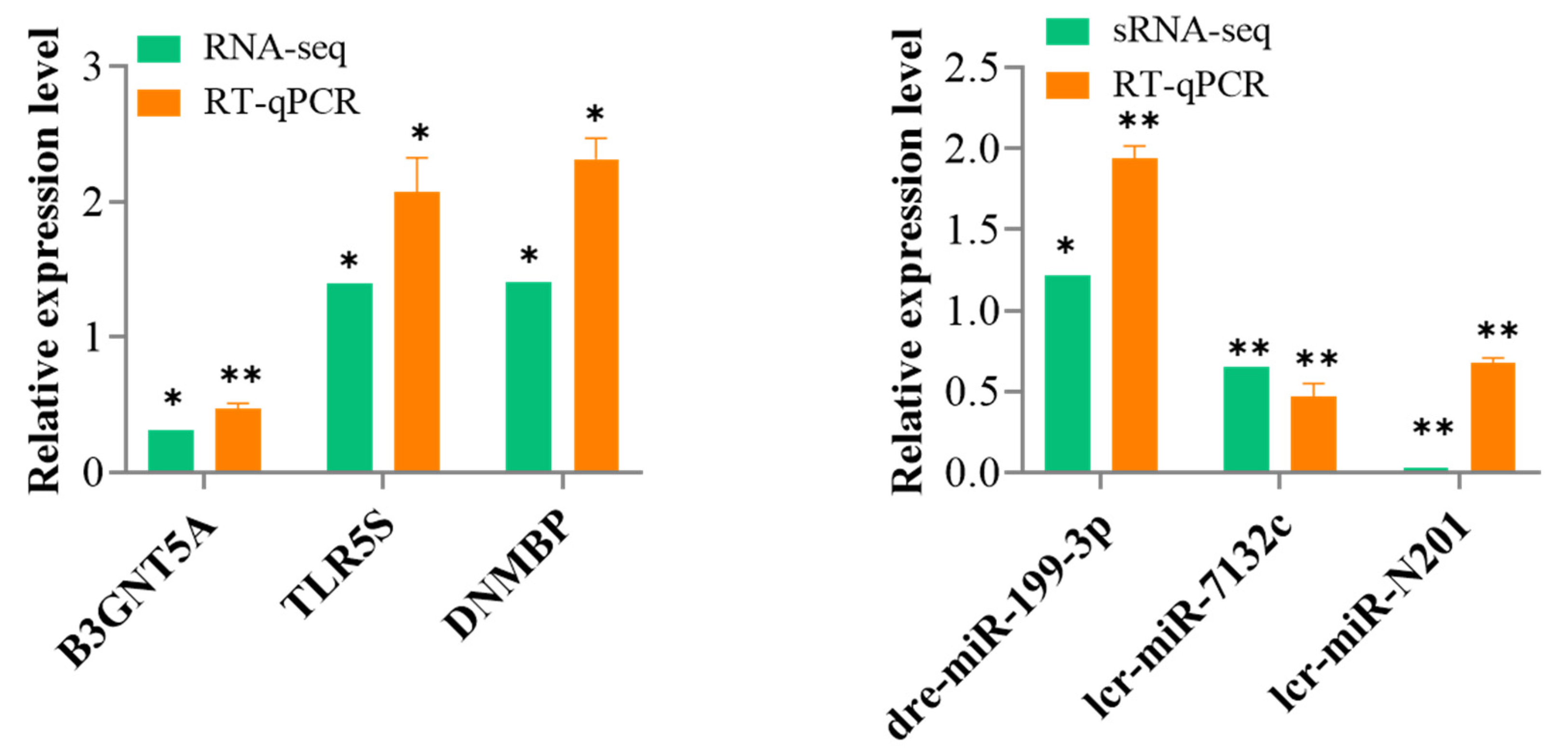
3.6. Validation of the DEmiRNAs and DEmRNAs Using RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Komoll, R.M.; Hu, Q.; Olarewaju, O.; von Döhlen, L.; Yuan, Q.; Xie, Y.; Tsay, H.C.; Daon, J.; Qin, R.; Manns, M.P.; et al. MicroRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. J. Hepatol. 2021, 74, 122–134. [Google Scholar] [CrossRef]
- Ito, Y.; Matsuzaki, T.; Ayabe, F.; Mokuda, S.; Kurimoto, R.; Matsushima, T.; Tabata, Y.; Inotsume, M.; Tsutsumi, H.; Liu, L.; et al. Both microRNA-455-5p and -3p repress hypoxia-inducible factor-2α expression and coordinately regulate cartilage homeostasis. Nat. Commun. 2021, 12, 4148. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, J.S.; Lee, S.; Lee, H.; Yoon, J.S.; Hong, J.H.; Chun, S.H.; Sun, S.; Won, H.S.; Hong, S.A.; et al. QKI, A miR-200 target gene, suppresses epithelial-to-mesenchymal transition and tumor growth. Int. J. Cancer 2019, 145, 1585–1595. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Leung, A.K.; Sharp, P.A. MicroRNA functions in stress responses. Mol. Cell 2010, 40, 205–215. [Google Scholar] [CrossRef]
- Lee, C.Q.E.; Kerouanton, B.; Chothani, S.; Zhang, S.; Chen, Y.; Mantri, C.K.; Hock, D.H.; Lim, R.; Nadkarni, R.; Huynh, V.T.; et al. Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. Nat. Commun. 2021, 12, 2130. [Google Scholar] [CrossRef]
- Bamunuarachchi, G.; Yang, X.; Huang, C.; Liang, Y.; Guo, Y.; Liu, L. MicroRNA-206 inhibits influenza A virus replication by targeting tankyrase 2. Cell. Microbiol. 2021, 23, e13281. [Google Scholar] [CrossRef]
- Chen, X.M.; Splinter, P.L.; O’Hara, S.P.; LaRusso, N.F. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J. Biol. Chem. 2007, 282, 28929–28938. [Google Scholar] [CrossRef]
- Ordas, A.; Kanwal, Z.; Lindenberg, V.; Rougeot, J.; Mink, M.; Spaink, H.P.; Meijer, A.H. MicroRNA-146 function in the innate immune transcriptome response of zebrafish embryos to Salmonella typhimurium infection. BMC Genom. 2013, 14, 696. [Google Scholar] [CrossRef]
- Ramberg, S.; Krasnov, A.; Colquhoun, D.; Wallace, C.; Andreassen, R. Expression analysis of Moritella viscosa-challenged Atlantic salmon identifies disease-responding genes, microRNAs and their predicted target genes and pathways. Int. J. Mol. Sci. 2022, 23, 11200. [Google Scholar] [CrossRef]
- Qiao, Y.; Mao, Y.; Wang, J.; Chen, R.; Libing, Z.; Su, Y.Q.; Chen, J.; Zheng, W.Q. Analysis of liver and gill miRNAs of Larimichthys crocea against Cryptocryon irritans challenge. Fish Shellfish Immunol. 2016, 59, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Dhanagovind, P.T.; Kujur, P.K.; Swain, R.K.; Banerjee, S. IL-6 signaling protects zebrafish larvae during Staphylococcus epidermidis infection in a bath immersion model. J. Immunol. 2021, 207, 2129–2142. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Abdellaoui, N.; Kim, K.H. Effect of miR-155 as a molecular adjuvant of DNA vaccine against VHSV in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2019, 88, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, T.D.; Nikapitiya, C.; Lee, J.; De Zoysa, M. Potential immune regulatory role of miR-146a upon Aeromonas hydrophila and Edwardsiella piscicida infections in zebrafish. Braz. J. Microbiol. 2020, 51, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, W.; Wang, B.; Zhang, S.; Xiao, Y.; Han, F.; Wang, Z. UBE2G1 is a critical component of immune response to the infection of Pseudomonas plecoglossicida in large yellow croaker (Larimichthys crocea). Int. J. Mol. Sci. 2022, 23, 8298. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Huo, J.; Guan, Y.; Fan, D.; Xiao, X.; Wei, J.; Li, Q.; Mu, P.; Ao, J.; Chen, X. An improved genome assembly for Larimichthys crocea reveals hepcidin gene expansion with diversified regulation and function. Commun. Biol. 2018, 1, 195. [Google Scholar] [CrossRef]
- Li, Q.; Chen, Y.; Xu, L.; Yang, Y.; Wen, Q.; Gu, L.; Ao, J.; Chen, X. Identification and bioactivity of a granulocyte colony-stimulating factor a homologue from large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. 2020, 98, 167–175. [Google Scholar] [CrossRef]
- Wang, L.; Sun, Y.; Zhao, L.; Xu, X.; Huang, L.; Qin, Y.; Su, Y.; Zhang, J.; Yan, Q. Dual RNA-seq uncovers the immune response of Larimichthys crocea to the secY gene of Pseudomonas plecoglossicida from the perspective of host-pathogen interactions. Fish Shellfish Immunol. 2019, 93, 949–957. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, L.; Li, C.; Shao, G.; Chen, X. Transcriptome analysis revealed multiple immune processes and energy metabolism pathways involved in the defense response of the large yellow croaker Larimichthys crocea against Pseudomonas plecoglossicida. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100886. [Google Scholar] [CrossRef]
- Ao, J.; Mu, Y.; Xiang, L.X.; Fan, D.; Feng, M.; Zhang, S.; Shi, Q.; Zhu, L.Y.; Li, T.; Ding, Y.; et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015, 11, e1005118. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Jin, D.; Yang, S.; Li, X.; Li, H.; Hu, S.; Sun, Y.; Yi, G.; Wang, P.; Rang, J.; et al. Pathogenicity of fish pathogen Pseudomonas plecoglossicida and preparation of its inactivated vaccine. Microb. Pathog. 2022, 166, 105488. [Google Scholar] [CrossRef] [PubMed]
- Zapata, A.; Diez, B.; Cejalvo, T.; Gutiérrez-de Frías, C.; Cortés, A. Ontogeny of the immune system of fish. Fish Shellfish Immunol. 2006, 20, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, R.; Høyheim, B. miRNAs associated with immune response in teleost fish. Dev. Comp. Immunol. 2017, 75, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Salinas, I.; Zhang, Y.A.; Sunyer, J.O. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 2011, 35, 1346–1365. [Google Scholar] [CrossRef]
- Ichiyama, K.; Dong, C. The role of miR-183 cluster in immunity. Cancer Lett. 2019, 443, 108–114. [Google Scholar] [CrossRef]
- Luo, C.; Xin, H.; Zhou, Z.; Hu, Z.; Sun, R.; Yao, N.; Sun, Q.; Borjigin, U.; Wu, X.; Fan, J.; et al. Tumor-derived exosomes induce immunosuppressive macrophages to foster intrahepatic cholangiocarcinoma progression. Hepatology 2022, 76, 982–999. [Google Scholar] [CrossRef]
- Trinh, T.L.; Kandell, W.M.; Donatelli, S.S.; Tu, N.; Tejera, M.M.; Gilvary, D.L.; Eksioglu, E.A.; Burnette, A.; Adams, W.A.; Liu, J.; et al. Immune evasion by TGFβ-induced miR-183 repression of MICA/B expression in human lung tumor cells. Oncoimmunology 2019, 8, e1557372. [Google Scholar] [CrossRef]
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar] [CrossRef]
- Muñoz-Flores, C.; Astuya, A.; Roa, F.J.; Romero, A.; Acosta, J.; Sánchez, O.; Toledo, J.R. Activation of membrane-bound and soluble Toll-like Receptors 5 in Salmo salar depends on the MyD88 signalling pathway. Biochim. Biophys. Acta. Gen. Subj. 2018, 1862, 2215–2225. [Google Scholar] [CrossRef]
- Umasuthan, N.; Bathige, S.; Thulasitha, W.S.; Jayasooriya, R.; Shin, Y.; Lee, J. Identification of a gene encoding a membrane-anchored toll-like receptor 5 (TLR5M) in Oplegnathus fasciatus that responds to flagellin challenge and activates NF-κB. Fish Shellfish Immunol. 2017, 62, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; Shen, Y.B.; Fu, J.J.; Yu, H.Y.; Huang, W.J.; Lu, L.Q.; Li, J.L. MicroRNA-induced negative regulation of TLR-5 in grass carp, Ctenopharyngodon idella. Sci. Rep. 2016, 6, 18595. [Google Scholar] [CrossRef] [PubMed]


| Sample | Raw Reads | Clean Reads | Mapped Reads | Q30 |
|---|---|---|---|---|
| PBS-12 h_1 | 14,873,980 | 13,430,011 (90.29%) | 9,965,946 (74.21%) | 98.86% |
| PBS-12 h_2 | 13,073,333 | 11,835,003 (90.53%) | 9,709,046 (82.04%) | 98.87% |
| PBS-12 h_3 | 15,481,958 | 13,709,443 (88.55%) | 10,968,433 (80.01%) | 98.88% |
| PBS-24 h_1 | 14,564,415 | 13,416,481 (92.12%) | 8,802,529 (65.61%) | 98.79% |
| PBS-24 h_2 | 13,325,699 | 11,955,087 (89.71%) | 9,225,816 (77.17%) | 98.75% |
| PBS-24 h_3 | 13,852,531 | 12,773,001 (92.21%) | 6,039,554 (47.28%) | 98.79% |
| PP-12 h_1 | 16,836,202 | 15,154,185 (90.01%) | 12,840,447 (84.73%) | 98.86% |
| PP-12 h_2 | 11,060,707 | 10,177,970 (92.02%) | 8,270,715 (81.26%) | 98.81% |
| PP-12 h_3 | 12,941,407 | 11,873,131 (91.75%) | 9,411,944 (79.27%) | 98.85% |
| PP-24 h_1 | 15,204,863 | 13,970,932 (91.88%) | 11,610,363 (83.10%) | 98.81% |
| PP-24 h_2 | 15,249,538 | 14,175,941 (92.96%) | 11,748,698 (82.88%) | 98.81% |
| PP-24 h_3 | 12,695,840 | 11,562,477 (91.07%) | 9,415,271 (81.43%) | 98.88% |
| Sample | Raw Reads | Clean Reads | Mapped Reads | Q30 |
|---|---|---|---|---|
| PBS-12 h_1 | 43,759,090 | 43,236,928 (98.81%) | 38,690,266 (89.48%) | 93.22% |
| PBS-12 h_2 | 47,559,984 | 46,977,842 (98.78%) | 41,848,130 (89.08%) | 93.04% |
| PBS-12 h_3 | 46,459,984 | 45,917,460 (98.83%) | 41,084,034 (89.47%) | 93.1% |
| PBS-24 h_1 | 48,368,716 | 47,851,156 (98.83%) | 43,120,810 (90.11%) | 93.49% |
| PBS-24 h_2 | 48,412,476 | 47,868,020 (98.88%) | 42,935,931 (89.7%) | 93.22% |
| PBS-24 h_3 | 50,151,150 | 49,586,402 (98.87%) | 44,435,722 (89.61%) | 93.33% |
| PP-12 h_1 | 47,578,048 | 47,059,414 (98.91%) | 42,137,958 (89.54%) | 93.43% |
| PP-12 h_2 | 45,091,328 | 44,557,844 (98.82%) | 39,893,631 (89.53%) | 93.17% |
| PP-12 h_3 | 50,707,652 | 50,156,332 (98.91%) | 44,954,501 (89.63%) | 93.44% |
| PP-24 h_1 | 46,781,528 | 46,222,554 (98.81%) | 41,453,383 (89.68%) | 93.04% |
| PP-24 h_2 | 46,402,592 | 45,886,162 (98.89%) | 41,196,858 (89.78%) | 93.43% |
| PP-24 h_3 | 52,820,076 | 52,241,174 (98.90%) | 46,817,324 (89.62%) | 93.38% |
| GO ID | Description | Number of DEmRNAs | p Value |
|---|---|---|---|
| GO terms significantly enriched in the DEmRNAs at 12 h post-injection | |||
| GO:0002274 | Interleukin-8 receptor binding | 3 | 0.000285 |
| GO:0032873 | CXCR chemokine receptor binding | 2 | 0.00031 |
| GO:0060326 | Lysosome | 5 | 0.001027 |
| GO:0005153 | Myeloid leukocyte activation | 2 | 0.001815 |
| GO:0045236 | Negative regulation of stress-activated MAPK cascade | 2 | 0.001815 |
| GO:0042119 | Cell chemotaxis | 2 | 0.001815 |
| GO:0036230 | Neutrophil activation | 2 | 0.001815 |
| GO:0030595 | Granulocyte activation | 4 | 0.003971 |
| GO:0051707 | Leukocyte chemotaxis | 11 | 0.005099 |
| GO:0043207 | Response to other organism | 11 | 0.005564 |
| GO:0009607 | Response to external biotic stimulus | 11 | 0.005727 |
| GO:0043409 | Response to biotic stimulus | 2 | 0.006134 |
| GO:0050900 | Negative regulation of MAPK cascade | 4 | 0.007411 |
| GO:0030593 | Leukocyte migration | 3 | 0.008357 |
| GO:0005764 | Neutrophil chemotaxis | 4 | 0.009344 |
| GO:1990266 | Neutrophil migration | 3 | 0.01046 |
| GO:0071621 | Granulocyte chemotaxis | 3 | 0.011616 |
| GO:0097530 | Granulocyte migration | 3 | 0.01414 |
| GO:0009617 | Response to bacterium | 6 | 0.016991 |
| GO:0002281 | Macrophage activation involved in immune response | 1 | 0.017613 |
| GO:0002366 | Leukocyte activation involved in immune response | 1 | 0.017613 |
| GO:0002263 | Cell activation involved in immune response | 1 | 0.017613 |
| GO:0002275 | Myeloid cell activation involved in immune response | 1 | 0.017613 |
| GO:0071466 | Cellular response to xenobiotic stimulus | 2 | 0.039336 |
| GO:0032526 | Response to retinoic acid | 2 | 0.039336 |
| GO:0045321 | Leukocyte activation | 3 | 0.042277 |
| GO:0031349 | Positive regulation of defense response | 3 | 0.042277 |
| GO:0006952 | Defense response | 9 | 0.046707 |
| GO terms significantly enriched in the DEmRNAs at 24 h post-injection | |||
| GO:0034138 | Toll-like receptor 3 signaling pathway | 2 | 0.005044 |
| GO:0032642 | Regulation of chemokine production | 2 | 0.005044 |
| GO:0032722 | Positive regulation of chemokine production | 2 | 0.005044 |
| GO:0006909 | Phagocytosis | 2 | 0.047425 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Zhang, Y.; Shao, G.; Chen, Y.; Shen, Y.; Mu, Y.; Chen, X. Comparative Transcriptomics Reveals the microRNA-Mediated Immune Response of Large Yellow Croaker (Larimichthys crocea) to Pseudomonas plecoglossicida Infection. Fishes 2023, 8, 10. https://doi.org/10.3390/fishes8010010
Chen H, Zhang Y, Shao G, Chen Y, Shen Y, Mu Y, Chen X. Comparative Transcriptomics Reveals the microRNA-Mediated Immune Response of Large Yellow Croaker (Larimichthys crocea) to Pseudomonas plecoglossicida Infection. Fishes. 2023; 8(1):10. https://doi.org/10.3390/fishes8010010
Chicago/Turabian StyleChen, Huazhi, Yameng Zhang, Guangming Shao, You Chen, Yibo Shen, Yinnan Mu, and Xinhua Chen. 2023. "Comparative Transcriptomics Reveals the microRNA-Mediated Immune Response of Large Yellow Croaker (Larimichthys crocea) to Pseudomonas plecoglossicida Infection" Fishes 8, no. 1: 10. https://doi.org/10.3390/fishes8010010
APA StyleChen, H., Zhang, Y., Shao, G., Chen, Y., Shen, Y., Mu, Y., & Chen, X. (2023). Comparative Transcriptomics Reveals the microRNA-Mediated Immune Response of Large Yellow Croaker (Larimichthys crocea) to Pseudomonas plecoglossicida Infection. Fishes, 8(1), 10. https://doi.org/10.3390/fishes8010010








