Cloning and Expression of Sox2 and Sox9 in Embryonic and Gonadal Development of Lutraria sieboldii
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue and Embryo Collection
2.2. Gene Isolation and Cloning
2.3. Bioinformatics Analysis
2.4. Tissue Distribution and LsSox2 and LsSox9 Expression Profiles in Adults and Embryos
2.5. Statistical Analysis
3. Results and Analysis
3.1. Cloning and Sequencing Analysis of the LsSox2 and LsSox9 Genes
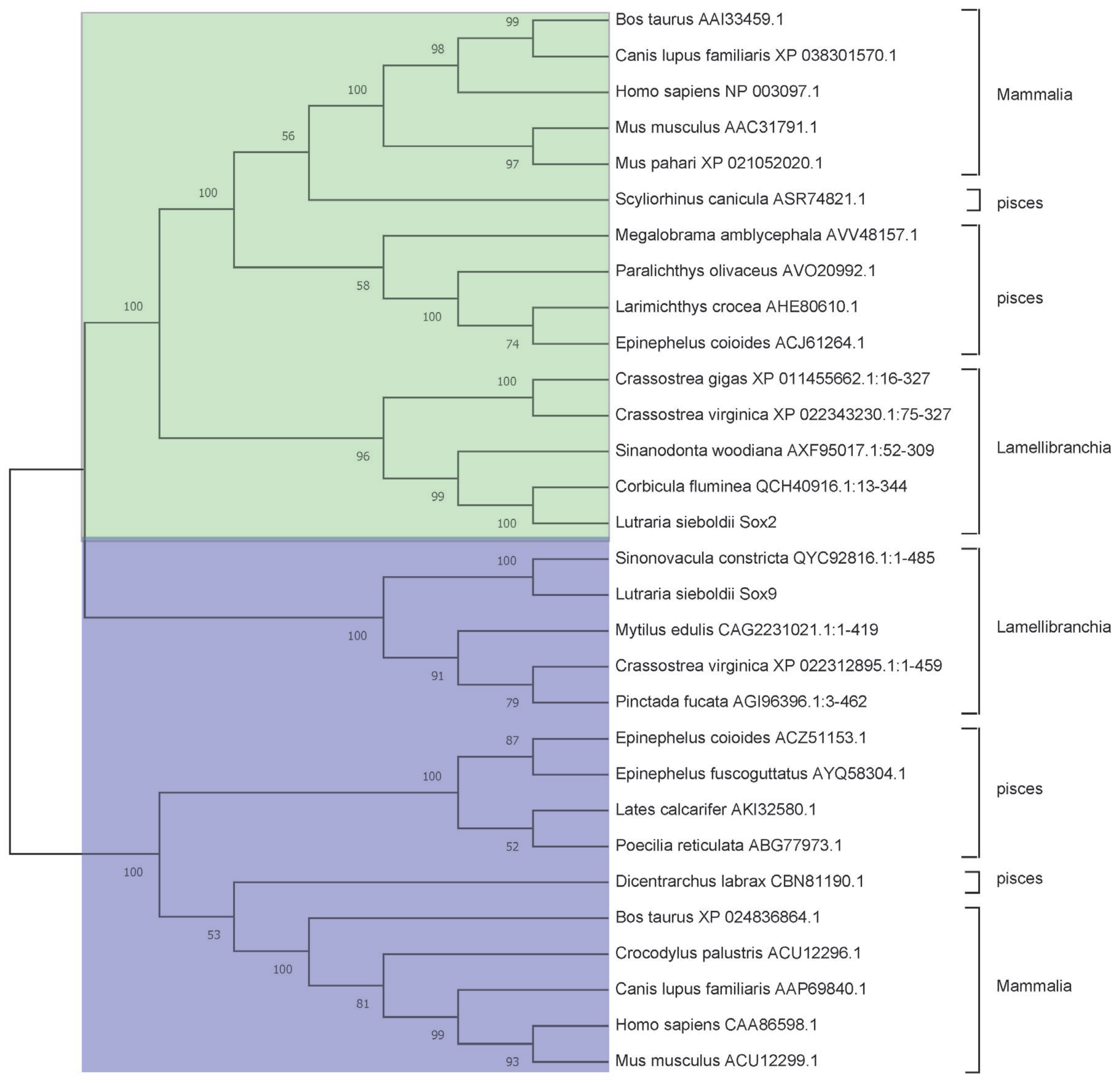
3.2. Phylogenetic Analysis
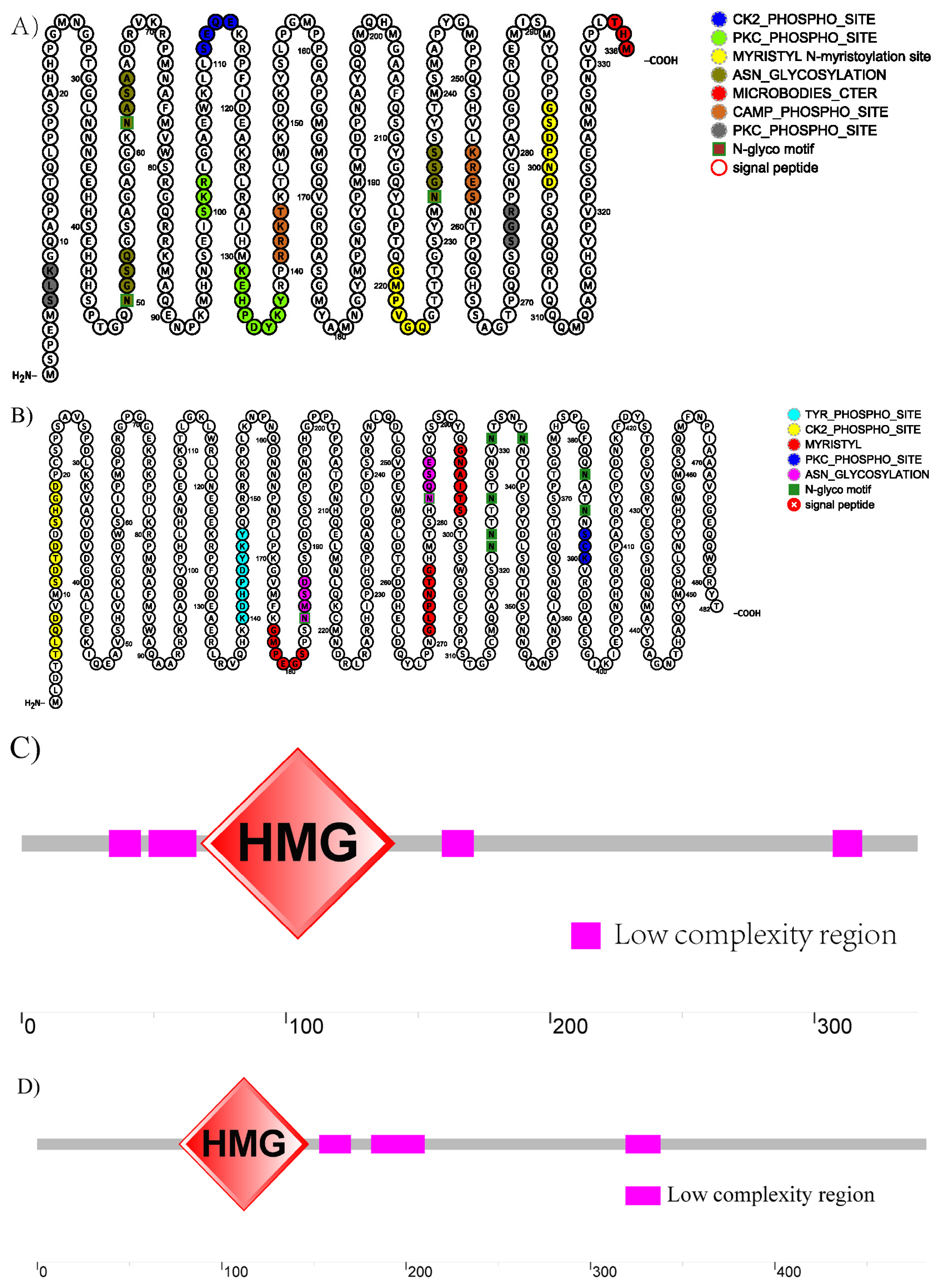
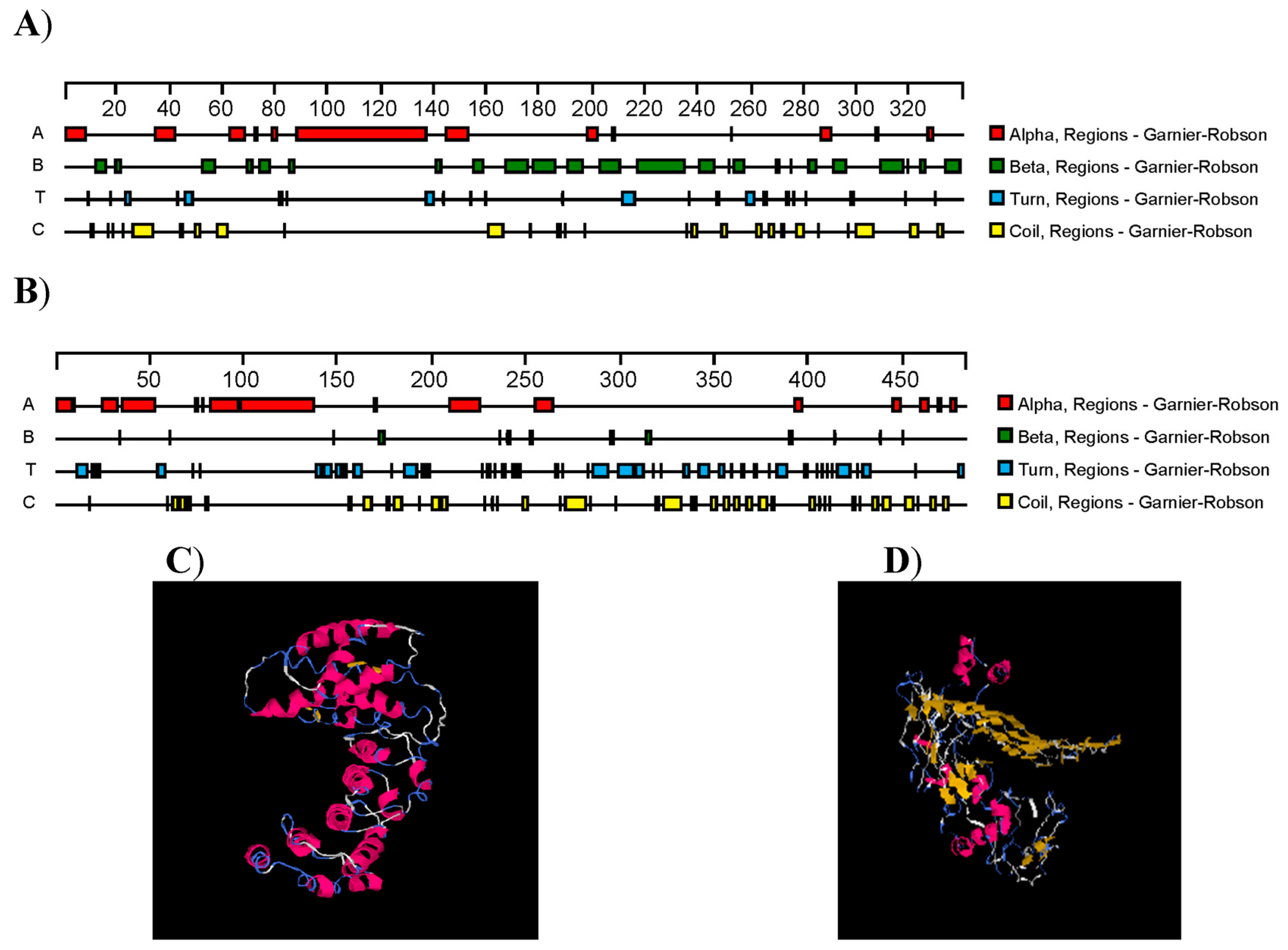
3.3. Bioinformatics Analysis
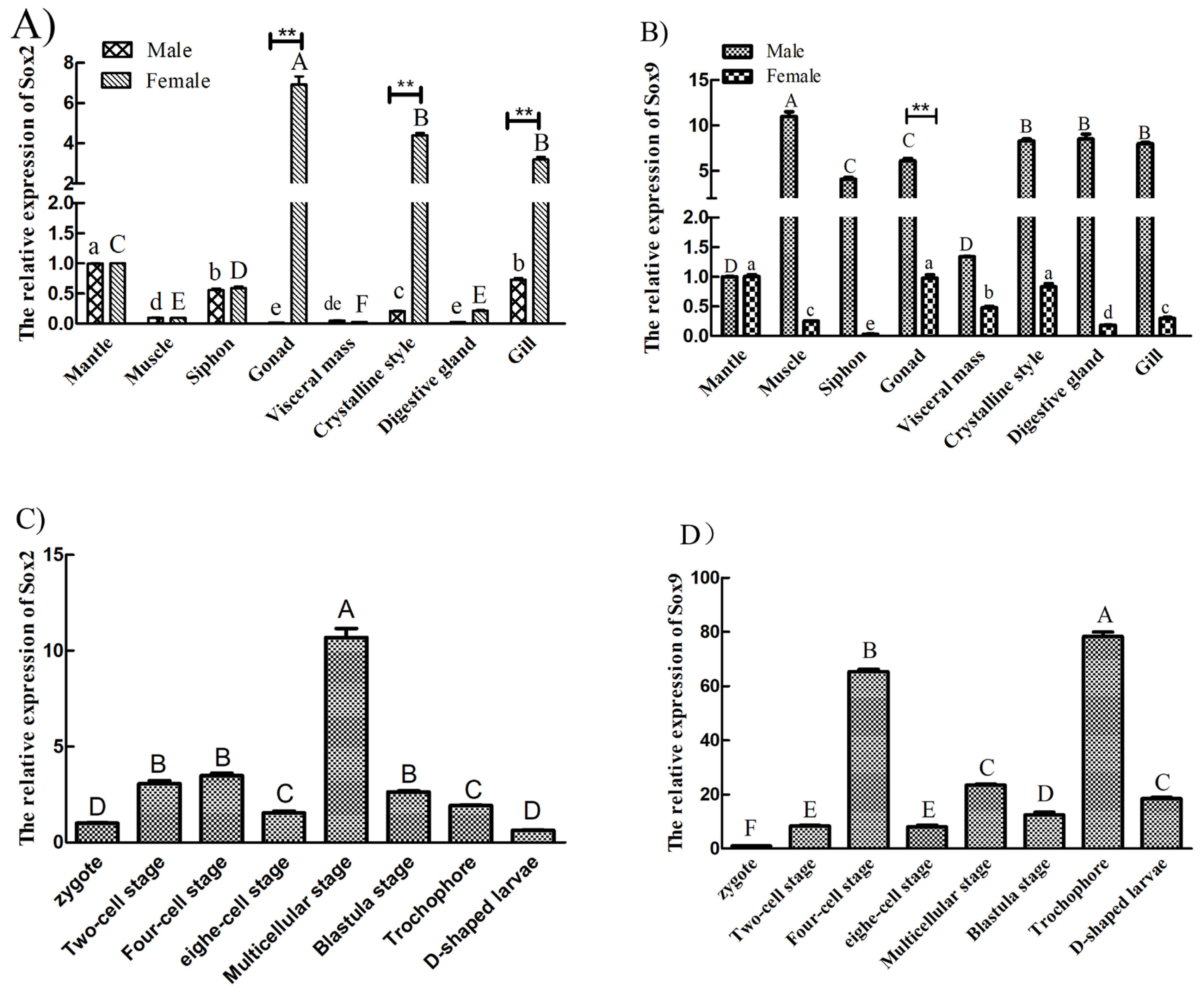
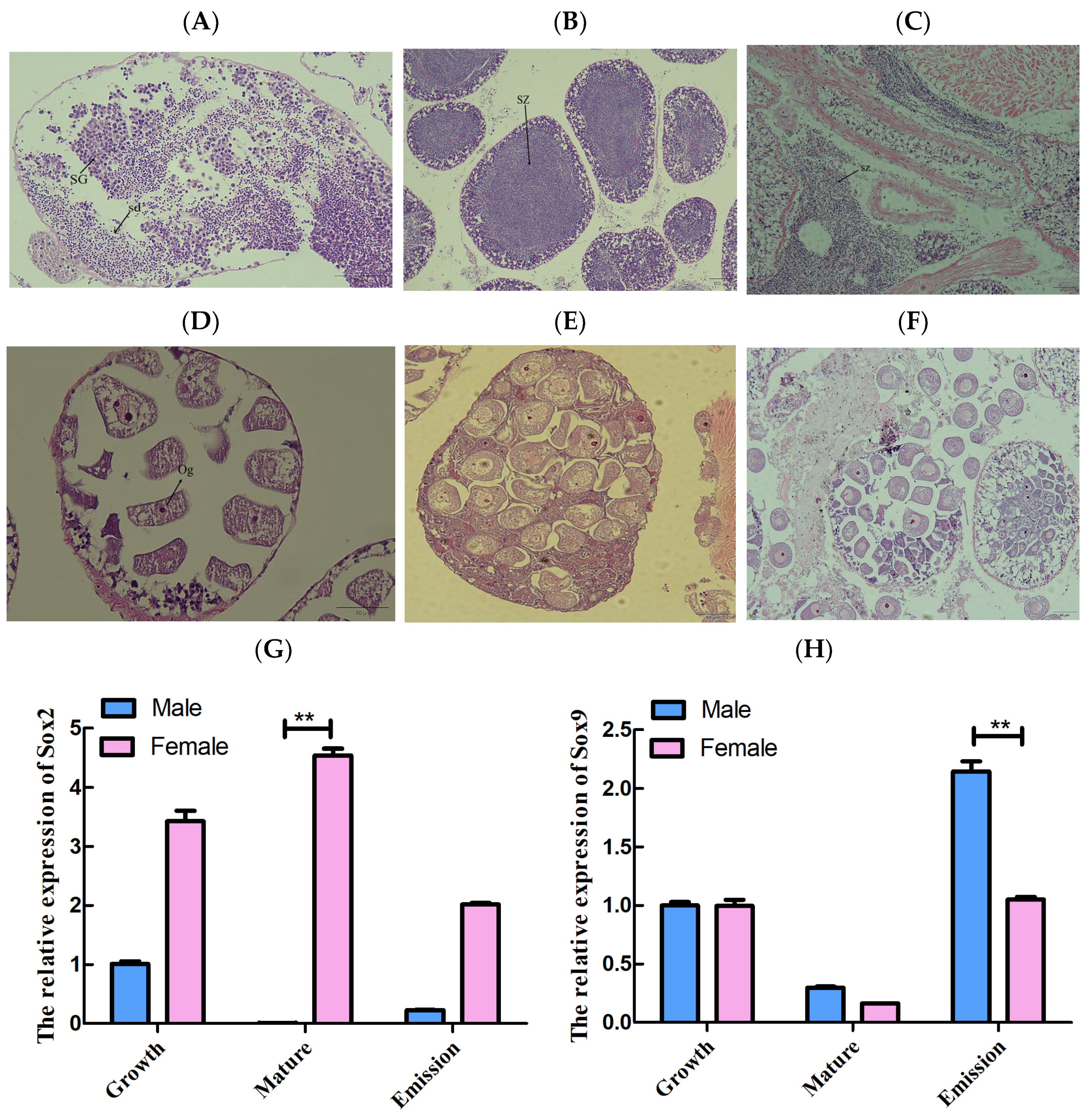
3.4. Tissue Distribution of LsSox2 and LsSox9 mRNA
3.5. Gene Expression Analysis of L. sieboldii Sox Genes in Gonads
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grimm, D.; Bauer, J.; Wise, P.; Krüger, M.; Simonsen, U.; Wehland, M.; Infanger, M.; Corydon, T.J. The role of SOX family members in solid tumours and metastasis. Semin. Cancer Biol. 2020, 67, 122–153. [Google Scholar] [CrossRef]
- She, Z.-Y.; Yang, W.-X. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015, 94, 547–563. [Google Scholar] [CrossRef]
- Schepers, G.E.; Teasdale, R.D.; Koopman, P. Twenty Pairs of Sox: Extent, Homology, and Nomenclature of the Mouse and Human Sox Transcription Factor Gene Families. Dev. Cell 2002, 3, 167–170. [Google Scholar] [CrossRef]
- Hou, L.; Srivastava, Y.; Jauch, R. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2017, 63, 2–12. [Google Scholar] [CrossRef]
- Kashimada, K.; Koopman, P. Sry: The master switch in mammalian sex determination. Development 2010, 137, 3921–3930. [Google Scholar] [CrossRef]
- Miyagi, S.; Kato, H.; Okuda, A. Role of SoxB1 transcription factors in development. Cell. Mol. Life Sci. 2009, 66, 3675. [Google Scholar] [CrossRef]
- Bernard, P.; Tang, P.; Liu, S.; Dewing, P.; Harley, V.R.; Vilain, E. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum. Mol. Genet. 2003, 12, 1755–1765. [Google Scholar] [CrossRef]
- Huang, X.; Wu, C.; Gong, K.; Chen, Q.; Gu, Q.; Qin, H.; Zhao, C.; Yu, T.; Yang, L.; Fu, W.; et al. Sox Gene Family Revealed Genetic Variations in Autotetraploid Carassius auratus. Front. Genet. 2020, 11, 804–804. [Google Scholar] [CrossRef]
- Fu, L.; Shi, Y.-B. The Sox transcriptional factors: Functions during intestinal development in vertebrates. Semin. Cell Dev. Biol. 2017, 63, 58–67. [Google Scholar] [CrossRef]
- Bowles, J.; Schepers, G.; Fau-Koopman, P.; Koopman, P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000, 227, 239–255. [Google Scholar] [CrossRef]
- Kuzmichev, A.N.; Kim Sk Fau-D’Alessio, A.C.; D’Alessio Ac Fau-Chenoweth, J.G.; Chenoweth Jg Fau-Wittko, I.M.; Wittko Im Fau-Campanati, L.; Campanati, L.; Fau-McKay, R.D.; McKay, R.D. Sox2 acts through Sox21 to regulate transcription in pluripotent and differentiated cells. Curr. Biol. 2012, 22, 1705–1710. [Google Scholar] [CrossRef] [PubMed]
- Archer, T.C.; Jin, J.; Casey, E.S. Interaction of Sox1, Sox2, Sox3 and Oct4 during primary neurogenesis. Dev. Biol. 2011, 350, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.A.; Hos, D.; Fau-Küspert, M.; Küspert, M.; Fau-Lang, R.A.; Lang Ra Fau-Lovell-Badge, R.; Lovell-Badge, R.; Fau-Wegner, M.; Wegner, M.; Fau-Reiprich, S.; et al. Stem cell factor Sox2 and its close relative Sox3 have differentiation functions in oligodendrocytes. Development 2014, 141, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, Z.; Shao, K.; Fan, L.; Yang, L.; Song, H.; Liu, M.; Wang, Z.; Wang, X.; Zhang, Q. Identification and characterization of a Sox2 homolog in the Japanese flounder Paralichthys olivaceus. Gene 2014, 544, 165–176. [Google Scholar] [CrossRef]
- Cui, C.-P.; Zhang, Y.; Wang, C.; Yuan, F.; Li, H.; Yao, Y.; Chen, Y.; Li, C.; Wei, W.; Liu, C.H.; et al. Dynamic ubiquitylation of Sox2 regulates proteostasis and governs neural progenitor cell differentiation. Nat. Commun. 2018, 9, 4648. [Google Scholar] [CrossRef]
- Xia, X.; Guan, C.; Chen, J.; Qiu, M.; Qi, J.; Wei, M.; Wang, X.; Zhang, K.; Lu, S.; Zhang, L.; et al. Molecular characterization of AwSox2 from bivalve Anodonta woodiana: Elucidating its player in the immune response. Innate Immun. 2020, 26, 381–397. [Google Scholar] [CrossRef]
- Wan, H.; Liao, J.; Zhang, Z.; Zeng, X.; Liang, K.; Wang, Y. Molecular cloning, characterization, and expression analysis of a sex-biased transcriptional factor sox9 gene of mud crab Scylla paramamosain. Gene 2021, 774, 145423. [Google Scholar] [CrossRef]
- Takamatsu, N.; Kanda, H.; Ito, M.; Yamashita, A.; Yamashita, S.; Shiba, T. Rainbow trout SOX9: cDNA cloning, gene structure and expression. Gene 1997, 202, 167–170. [Google Scholar] [CrossRef]
- Du, Q.Y.; Wang, F.Y.; Hua, H.Y.; Chang, Z.J. Cloning and study of adult-tissue-specific expression of Sox9 in Cyprinus carpio. J. Genet. 2007, 86, 85–91. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, K.; Ren, T.; Zhang, Z.; Ma, Z.; Dan, S.F.; Lan, Z.; Lu, M.; Fang, H.; Zhang, Y.; et al. The characterization, expression and activity analysis of three superoxide dismutases in Eriocheir hepuensis under azadirachtin stress. Fish. Shellfish. Immunol. 2021, 117, 228–239. [Google Scholar] [CrossRef]
- Lu, M.; Yang, J.; Wang, Z.; Song, J.; Hu, Y.; Wang, P.; Zhang, H.; Xu, Y.; Zhu, P. Cloning and expression of the ChGstα and ChGstκ genes in the gills of Crassostrea hongkongensis under nanoparticulate and ionic Zn stress. Comp. Biochem. Physiology. Toxicol. Pharmacol. CBP 2021, 244, 109007. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Pan, H.; Lu, M.; Liu, Q.; Shafique, L.; Peng, J.; Ahmed, T.; Wang, R.; Zhang, H.; Wang, Q.; et al. Analysis of transcripts and splice isoforms in red claw crayfish (Cherax quadricarinatus) using single-molecule long-read sequencing. Aquaculture 2021, 541, 736828. [Google Scholar] [CrossRef]
- Jiang, Y.-H.; Han, K.-H.; Wang, S.-H.; Chen, Y.; Wang, Y.-L.; Zhang, Z.-P. Identification and expression of transcription factor sox2 in large yellow croaker Larimichthys crocea. Theriogenology 2018, 120, 123–137. [Google Scholar] [CrossRef]
- Kurtz, S.; Lucas-Hahn, A.; Schlegelberger, B.; Göhring, G.; Niemann, H.; Mettenleiter, T.C.; Petersen, B. Knockout of the HMG domain of the porcine SRY gene causes sex reversal in gene-edited pigs. Proc. Natl. Acad. Sci. USA 2021, 118, e2008743118. [Google Scholar] [CrossRef]
- Jeng, S.R.; Wu, G.C.; Yueh, W.S.; Kuo, S.F.; Dufour, S.; Chang, C.F. Gonadal development and expression of sex-specific genes during sex differentiation in the Japanese eel. Gen. Comp. Endocrinol. 2018, 257, 74–85. [Google Scholar] [CrossRef]
- Kanda, H.; Kojima, M.; Miyamoto, N.; Ito, M.; Takamatsu, N.; Yamashita, S.; Shiba, T. Rainbow trout Sox24, a novel member of the Sox family, is a transcriptional regulator during oogenesis. Gene 1998, 211, 251–257. [Google Scholar] [CrossRef]
- Kamachi, Y.; Uchikawa, M.; Kondoh, H. Pairing SOX off: With partners in the regulation of embryonic development. Trends Genet. TIG 2000, 16, 182–187. [Google Scholar] [CrossRef]
- Adolfi, M.C.; Carreira, A.C.; Jesus, L.W.; Bogerd, J.; Funes, R.M.; Schartl, M.; Sogayar, M.C.; Borella, M.I. Molecular cloning and expression analysis of dmrt1 and sox9 during gonad development and male reproductive cycle in the lambari fish, Astyanax altiparanae. Reprod. Biol. Endocrinol. RBE 2015, 13, 2. [Google Scholar] [CrossRef]
- Vining, B.; Ming, Z.; Bagheri-Fam, S.; Harley, V. Diverse Regulation but Conserved Function: SOX9 in Vertebrate Sex Determination. Genes 2021, 12, 486. [Google Scholar] [CrossRef]
- Raghuveer, K.; Senthilkumaran, B. Isolation of sox9 duplicates in catfish: Localization, differential expression pattern during gonadal development and recrudescence, and hCG-induced up-regulation of sox9 in testicular slices. Reproduction 2010, 140, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Degani, G. Expression of SOX3 and SOX9 Genes in Gonads of Blue Gourami. Adv. Biol. Chem. 2014, 4, 2162–2183. [Google Scholar] [CrossRef]
- Okuda, Y.; Ogura, E.; Kondoh, H.; Kamachi, Y. B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PLoS Genet. 2010, 6, e1000936. [Google Scholar] [CrossRef]
- White, M.D.; Angiolini, J.F.; Alvarez, Y.D.; Kaur, G.; Zhao, Z.W.; Mocskos, E.; Bruno, L.; Bissiere, S.; Levi, V.; Plachta, N. Long-Lived Binding of Sox2 to DNA Predicts Cell Fate in the Four-Cell Mouse Embryo. Cell 2016, 165, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Schultz, R.M. Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol. Reprod. 2011, 85, 409–416. [Google Scholar] [CrossRef]
- Lee, M.; Choi, K.H.; Oh, J.N.; Kim, S.H.; Lee, D.K.; Choe, G.C.; Jeong, J.; Lee, C.K. SOX2 plays a crucial role in cell proliferation and lineage segregation during porcine pre-implantation embryo development. Cell Prolif. 2021, 54, e13097. [Google Scholar] [CrossRef]
- Hu, Y.; Jin, S.; Fu, H.; Qiao, H.; Zhang, W.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Functional analysis of a SoxE gene in the oriental freshwater prawn, Macrobrachium nipponense by molecular cloning, expression pattern analysis, and in situ hybridization (de Haan, 1849). 3 Biotech. 2019, 10, 10. [Google Scholar] [CrossRef]

| Primers | Sequence (5′-3′) | Amplified Length (bp) | Tm (°C) | Note |
|---|---|---|---|---|
| LsSox2F1 | CAAGCACTGTTTTTGAGAGG | 1574 | 56 | CDS cloning |
| LsSox2R1 | TTCATTTCACGATTTAACGG | |||
| LsSox2dlF1 | TTTACCCGTTAAATCGTGAAAT | 92 | 59 | qRT-PCR |
| LsSox2dlR1 | TGAAAGTTGAGAATCGCCTCT | |||
| LsSox9F1 | AGCGAACTTGTTTGGACATA | 1504 | 57 | CDS cloning |
| LsSox9R1 | ACCGTTTCTCAGCGTCTAAT | |||
| LsSox9dlF1 | AAAGTAAGAGACGACGCAG | 123 | 58 | qRT-PCR |
| LsSox9dlR1 | GTAACGAGACACAGAGGGG | |||
| LsEf1dlF1 | CTGTCTTAGATTGTCACACTGCC | 104 | 65 | qRT-PCR |
| LsEf1dlR1 | CGTCTTAGGGTTGTCTTCCAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, M.; Xing, Z.; Zhou, Y.; Xu, Y.; Peng, H.; Zou, J.; Dan, S.F.; She, Z.; Wang, P.; Liu, J.; et al. Cloning and Expression of Sox2 and Sox9 in Embryonic and Gonadal Development of Lutraria sieboldii. Fishes 2022, 7, 392. https://doi.org/10.3390/fishes7060392
Lu M, Xing Z, Zhou Y, Xu Y, Peng H, Zou J, Dan SF, She Z, Wang P, Liu J, et al. Cloning and Expression of Sox2 and Sox9 in Embryonic and Gonadal Development of Lutraria sieboldii. Fishes. 2022; 7(6):392. https://doi.org/10.3390/fishes7060392
Chicago/Turabian StyleLu, Min, Zenghou Xing, Yurui Zhou, Youhou Xu, Huijing Peng, Jie Zou, Solomon Felix Dan, Zhicai She, Pengliang Wang, Jinfeng Liu, and et al. 2022. "Cloning and Expression of Sox2 and Sox9 in Embryonic and Gonadal Development of Lutraria sieboldii" Fishes 7, no. 6: 392. https://doi.org/10.3390/fishes7060392
APA StyleLu, M., Xing, Z., Zhou, Y., Xu, Y., Peng, H., Zou, J., Dan, S. F., She, Z., Wang, P., Liu, J., Qin, S., Yang, J., & Zhu, P. (2022). Cloning and Expression of Sox2 and Sox9 in Embryonic and Gonadal Development of Lutraria sieboldii. Fishes, 7(6), 392. https://doi.org/10.3390/fishes7060392






