Abstract
Genetically improved farmed tilapia (Oreochromis niloticus, GIFT) is prone to hepatic metabolic imbalances and fatty liver disease during intensive farming. Long non-coding RNAs (lncRNAs) perform essential roles in various biological processes, including lipid metabolism. However, the lncRNAs involved in hepatic lipid metabolism in tilapia have not yet been identified. In this study, Illumina sequencing and bioinformatic analyses were performed on the liver of juvenile male GIFT fed a high-fat diet (HFD, 18.5% lipid) or a normal-fat diet (NFD, 8% lipid) for 56 days. RNA-seq analyses revealed 299 differentially expressed (DE)-mRNAs and 284 DE-lncRNAs between these two groups. The transcript profiles of 14 candidates (seven DE-mRNA and seven DE-lncRNAs) were verified by qRT-PCR, and the results were consistent with the RNA-seq results. Furthermore, 65 cis target genes and 3610 trans target genes of DE-lncRNAs were predicted. Functional analyses suggested that multiple metabolic pathways are affected by a high fat intake, including the PPAR signaling, fatty acid degradation, and fatty acid metabolism pathways. A co-expression network analysis indicated that many lncRNAs interact with numerous genes involved in lipid metabolism, and that some genes are regulated by multiple lncRNAs. The expression patterns of three lncRNAs (MSTRG.14598.1, MSTRG.6725.3, and MSTRG.13364.2) and their potential target genes (faldh, slc25a48, and fabp7a) in the PPAR signaling pathway were investigated. Our study provides new information about lncRNAs associated with lipid metabolism in tilapia.
1. Introduction
Genetically improved farmed tilapia (GIFT, Oreochromis niloticus) is a widely farmed fish in China, especially in southern China, because of its rapid growth and excellent stress resistance [1]. However, fatty liver disease, which results from an imbalance of hepatic lipid metabolism, often occurs in intensively farmed tilapia [2]. There are many causes of fatty liver disease in fish, including nutritional imbalances, environmental stress, and various physiological disorders [3]. Among them, imbalanced nutrients in feed is the main cause of fatty liver disease in farmed tilapia [4,5]. Lipids, as a non-protein nutrient, are essential for fish. High-fat feeds are widely used in fish farming because they provide energy while saving feed protein [6,7]. However, an inappropriately high lipid intake can slow the growth of tilapia and reduce its stress resistance [5,8], thereby causing economic losses to the tilapia industry.
Lipid metabolism is an intricate biological process. Genes identified as crucial regulators of lipid metabolism include those encoding fatty acid synthetase (FAS) [9], peroxisome proliferator-activated receptor (PPAR) [10], and stearoyl-coa desaturase [11]. In our previous studies, we found that some non-coding RNAs, microRNA (miR)-34a, miR-205-5p, and miR-23a-3p, participate in the regulation of lipid metabolism in tilapia [4]. However, long non-coding RNAs (lncRNAs) have been rarely studied in tilapia, especially in terms of their involvement in lipid metabolism.
LncRNAs are RNAs more than 200 nucleotides (nt) in length and lack protein-coding ability. They are widely found in animals and plants [12]. Most of them are functional and regulate gene expression at epigenetic, transcriptional, and post-transcriptional levels [13,14,15]. Many studies have demonstrated that lncRNAs are key regulators of lipid metabolism. For example, LncNEAT1 was found to promote the expression of ACC, encoding acetyl-CoA carboxylase, and FAS, encoding fatty acid synthase, in hepatocytes by activating the mTOR/S6K1 signaling pathway. Interference with LncNEAT1 lentivirus reduced hepatic fat deposition in rats [16]. A study on human hepatic cells showed that LncHR1 affects fat synthesis by regulating the expression of the gene encoding sterol regulatory element binding protein-1c [17]. In mice, depletion of a liver-specific lncRNA (lncLSTR) was found to enhance the transcript levels of apoC2, leading to increased lipoprotein lipase activity and decreased serum triglyceride content [18]. Lipid metabolism-related lncRNAs have been identified in several fishes, including Chinese tongue sole [19] and zebrafish [20]. However, the lncRNAs associated with hepatic lipid metabolism in tilapia have not yet been explored. Therefore, to explore their functions in more detail, it is important to first identify and characterize them.
Previously, we reported a model system wherein fatty liver disease can be induced in juvenile male GIFT by a high-fat diet (HFD, 18.5% w/v lipid). Using this system, we compared metabolites between fish in a HFD group and those fed a normal-fat diet (NFD, 8% w/v lipid) [21]. Based on that study, hepatic profiles of mRNAs and lncRNAs were determined after 56 days of a HFD or NFD by RNA-seq. Then, bioinformatics techniques were conducted to predict the target mRNAs of the lncRNAs. Quantitative real-time PCR (qRT-PCR) was used to further analyze and investigate the expression patterns of several key lncRNAs involved in lipid metabolism and their target mRNAs on days 14, 35, and 56 of a HFD. Our findings provide valuable information for further analyses of the functional roles of lncRNAs in fat metabolism of tilapia.
2. Materials and Methods
2.1. Experimental Design and Sampling
Healthy male GIFT juveniles, which were purchased from the YangZhong Research Station (Yangzhong, China), were stocked in 600-L aerated water tanks (27–29 °C and pH 7.4–7.6) for 1 week. Fish were fed a commercial diet (Tongwei, China) two times per day during this 1-week acclimation period.
After acclimation, 180 healthy fish (average weight, 5.02 ± 0.01 g) were randomly divided into NFD (8% w/v lipid) and HFD (18.5% w/v lipid) groups in triplicate. Each tank housed 30 fish (n = 90 per group). The NFD and HFD were formulated as previously described [4]. The fish were fed to apparent satiation two times per day. The feed formulae are listed in Table S1.
The feeding trial lasted for 56 days. On days 14, 35, and 56, three fish from each tank were randomly caught and anaesthetized with 100 mg/L MS-222. Liver tissues collected from the sampled fish were frozen in liquid nitrogen and then stored at −80 °C until further molecular experiments. On day 56, liver tissue from another GIFT from each tank was collected for histological analyses. The use of fish in these experiments was approved by the Bioethical Committee of Freshwater Fisheries Research Center (2013863BCE) and all experimental manipulations complied with animal welfare requirements.
2.2. Hepatic Histological Analysis
The samples were fixed in 4% paraformaldehyde for 24 h and then dehydrated overnight in a 30% sucrose solution at 4 °C. Each sample was embedded in OCT compound and cut into 8-μm sections using a freezing microtome (Leica 3050S, Wetzlar, Germany). After rewarming to room temperature, the slices were immersed in oil red O staining solution for 10 min, immersed in 60% isopropyl alcohol for 30 s to remove the background color, and cleaned with pure water. After redyeing with Mayer’s hematoxylin for 3 min, the slices were sealed with glycerin gelatin and observed under a CX31 microscope (Olympus, Tokyo, Japan). The area of lipid droplets in six digital fields per group was analyzed using Image-Pro plus v6.0.0.260 [22].
2.3. RNA Extraction and Sequencing
Total RNA was isolated from liver tissues sampled at day 56 using TRIZOL reagent (Invitrogen, Carlsbad, USA) and genomic DNA was removed by DNase I (Promega, Madison, USA). RNA quality was assessed using a Bioanalyzer 2100 (Agilent, Palo Alto, USA) [23]. The sequencing was performed with three bio-replicates per group (RNA from three samples per group was mixed). Thus, six cDNA libraries (HFD_1, HFD_2, HFD_3, NFD_1, NFD_2, NFD_3) were constructed using an mRNA-Seq sample preparation kit (Illumina, San Diego, USA) after rRNA removal with the Epicentre Ribo-Zero Gold Kit (Illumina), following the manufacturer’s instructions. The library was sequenced on the Illumina Novaseq™ 6000 platform.
2.4. Data Filtering and lncRNA Identification
Raw reads were trimmed by Cutadapt v1.10 to eliminate low-quality reads and adaptor sequences. Sequence quality was checked by FastQC v0.10.1. HISAT2 v2.0.4 was used to map reads to the Nile tilapia genome (https://www.ncbi.nlm.nih.gov/genome/?term=Oreochromis+niloticus), accessed on 20 July 2018 [24]. The alignment results were then assembled into transcripts using StringTie v1.3.0 and classified with Gffcompare v.0.11.2. Among them, transcripts with the class code J, I, O, U, or X were further screened to identify lncRNAs. Among them, transcripts with the class code J, I, O, U, or X were further screened to identify lncRNAs.
To identify lncRNAs, we removed the following sequences: (1) those shorter than 200 bp or containing less than one exon; (2) those with reads coverage <3; and (3) those with known non-lncRNA annotations. Then, Coding Potential Calculator (CPC) v0.9-r2 [25] and Coding-Non-Coding Index (CNCI) v2.0 [26] were used to estimate coding potential of each remaining transcript. Transcripts with CPC score < 0 and CNCI score < 0 were considered to be lncRNAs [27].
2.5. Differential Expression Analyses of lncRNAs and mRNAs
StringTie v1.3.0 was used to standardize the expression levels of lncRNA and mRNA in each sample by calculating the FPKM (fragments per kilobase per million mapped reads) values. Then, EdgeR v3.22.3 was used to identify the differentially expressed (DE)-mRNAs and DE-lncRNAs between the HFD and NFD groups [28]. The criteria for differential expression between the two groups were |log2 fold change| > 1 and p < 0.05 [29].
2.6. Target Gene Prediction
Next, we predicted the potential cis and trans targets of DE-lncRNAs. Cis-acting lncRNAs target neighboring genes. In this study, genes located within 100 kb upstream or downstream of the de-lncRNA were selected as cis-regulated genes using Bedtools v2.17.0 [30]. LncRNAs can also regulate genes across chromosomes in a trans-acting manner. This interaction mainly depends on free energy demands for the formation of a secondary structure between the lncRNA and the target mRNA. Risearch v2.0 was used to investigate the relationships between lncRNAs and mRNAs [31]. The thresholds for screening trans-target genes were as follows: ≥10 directly interacting bases, and minimum free energy < −50 [32].
2.7. Pathway Enrichment Analysis
To explore the function of De-lncRNAs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted for the cis- and trans-target genes. The threshold for identifying significantly enriched GO terms and KEGG pathways was p < 0.05.
2.8. qRT-PCR Analyses
Total RNA was extracted from liver tissues sampled at days 14, 35, and 56 using the method described in 2.3. The cDNA was synthesized using PrimeScript RT Master Mix (Takara, Dalian, China) according to the manufacturer’s instructions. qPCR was performed on an ABI QuantStudio 5 instrument (ABI, Foster City, USA) with a SYBR® Premix Ex Taq kit (Takara). The reaction mixture (25 μL) contained 12.5 μL SYBR Premix Ex Taq II (2×), 0.5 μL ROX Dye II, 1 μL forward and reverse primers (10 μM), 2 μL cDNA template, and 8 μL RNase-free water. All reactions were performed in triplicate with the following thermal cycling program: 95 °C, for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Elongation factor 1α (ef1α) and β-actin were used as reference genes. Relative gene transcript levels were calculated by the 2−ΔΔCt method and ΔCt = Cttarget − (CtEF1α + Ctβ-actin)/2. The primer sequences are presented in Table 1. Data were expressed as the mean ± standard error and were analyzed using SPSS 22.0 (SPSS Inc., Chicago, USA). Significance (p < 0.05) was detected by one-way ANOVA followed by Duncan’s test.

Table 1.
Primer sequences used for qRT-PCR of DE-mRNAs and DE-lncRNAs.
3. Results
3.1. Hepatic Histological Analysis
Hepatic lipid deposition was observed by oil red O staining (Figure 1). Compared with the liver of fish from the NFD group (Figure 1B), the liver of fish from the HFD group had a much larger area of lipid droplets (Figure 1A, Table 2). This extensive lipid accumulation may have impaired normal liver function.
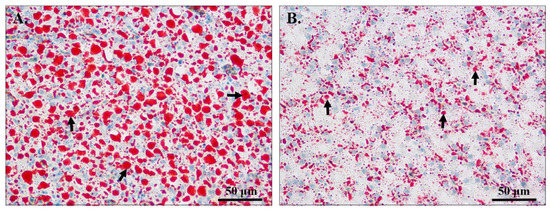
Figure 1.
Oil red O staining of liver tissues in genetically improved farmed tilapia (GIFT) fed a high-fat diet (HFD) or normal-fat diet (NFD) for 56 days. Bar = 50 μm. (A) Liver status of GIFT in the HFD group; (B) Liver status of GIFT in the NFD group. Black arrows indicate lipid droplets.

Table 2.
Area of lipid droplets in the liver of GIFT fed a HFD or NFD for 56 days.
3.2. Overview of Liver RNA Sequencing
Liver tissues from fish fed a HFD or NFD for 56 days were used to construct cDNA libraries for sequencing. The number of clean reads in each library ranged from 60,134,656 to 86,665,870 after filtering. The Q30 values of these libraries ranged from 96.77% to 97.91% (Table S2). Comparisons of the expression density of all transcripts and overall gene transcript levels among the six libraries (Figure S1A,B) confirmed the reliability of the Illumina sequencing data. For the HFD_1, HFD_2, HFD_3, NFD_1, NFD_2, and NFD_3 libraries, 87.11%, 86.96%, 87.10%, 87.32%, 86.80%, and 87.43% of the reads were mapped to the Nile tilapia genome, respectively (Table S2).
3.3. Identification and Quantification of lncRNAs
Several filtering steps were performed to identify lncRNAs from total transcripts, yielding 19,928 candidate lncRNAs. The full length of most lncRNAs ranged from 200 to 700 bp, while most full-length mRNAs were longer than 1000 bp (Figure 2A). A large number of lncRNAs contained only one or two exons (95.38%), while the number of exons in mRNAs was more variable (Figure 2B). The candidate lncRNAs were classified into five types (U, I, X, J, and O) by Gffcompare software. Most lncRNAs belonged to the U (intergenic) and I (intronic) classes, accounting for 65.11% and 25.45% of total lncRNAs, respectively (Figure 2C).
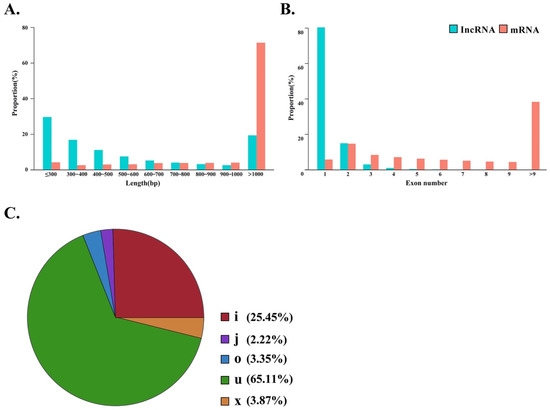
Figure 2.
Comparison of lncRNAs and mRNAs in GIFT. (A) Length distribution of mRNAs and lncRNAs. (B) Number of exons in mRNAs and lncRNAs. (C) Pie chart of lncRNA classification groups.
3.4. Screening of DE-lncRNAs and DE-mRNAs in Response to a High-Fat Diet
After 56 days of a HFD, 299 mRNAs and 284 lncRNAs in the GIFT liver showed significant changes in abundance (Table S3), including 137 up-regulated and 162 down-regulated mRNAs (Figure 3A), and 155 up-regulated and 129 down-regulated lncRNAs (Figure 3B). Radar plots were constructed to show the top 20 DE-mRNAs (nine up-regulated and 11 down-regulated mRNAs) (Figure 3C) and the top 20 DE-lncRNAs (13 up-regulated and seven down-regulated lncRNAs) (Figure 3D).
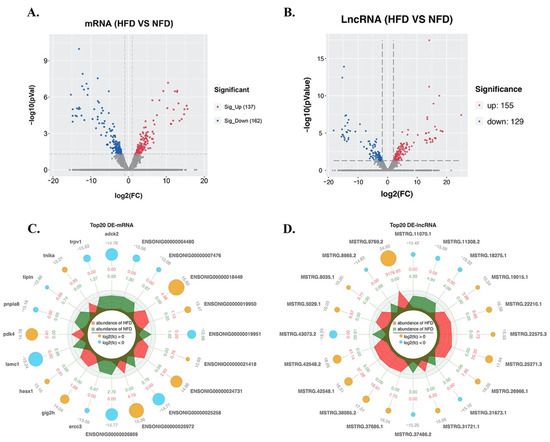
Figure 3.
Differential expression analysis of lncRNAs and mRNAs. Volcano plot of differentially expressed (DE)-mRNAs (A) and -lncRNAs (B); Radar plots of the top 20 DE-mRNAs (C) and DE-lncRNAs (D).
3.5. Functional Enrichment Analysis of DE-mRNAs
A GO enrichment analysis was conducted to investigate the biological functions of DE-mRNAs after 56 days of a HFD (Figure 4A and Figure S2). In the biological process category, DE-mRNAs were mainly enriched in the “cellular process”, “single-organism process” and “metabolic process” subcategories, among which “DNA topological change” and “membrane docking” were the most significantly enriched terms. In the cellular component category, DE-mRNAs were mainly enriched in the “cell”, “cell part”, and “organelle” subcategories, among which “fibrinogen complex” and “extracellular region” were the most significantly enriched terms. In the molecular function category, DE-mRNAs were mainly enriched in the “binding” and “catalytic activity” subcategories, with the top two GO terms being “glucagon receptor binding” and “peroxisome matrix targeting signal-1 binding”.
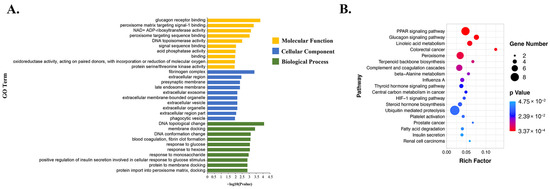
Figure 4.
GO and KEGG enrichment analysis of DE-mRNAs in the liver of the HFD group compared with the NFD group. (A) Top 10 GO terms in three categories: biological process, molecular function, and cellular component. (B) Top 20 enriched KEGG pathways.
The DE-mRNAs were further annotated at the KEGG database to identify crucial signaling pathways (Figure 4B). The top 20 KEGG pathways enriched with DE-mRNAs included glycolipid metabolism-related pathways, such as “PPAR signaling pathway”, “glucagon signaling pathway”, and “linoleic acid metabolism”.
3.6. Data Confirmation by qRT-PCR
In order to evaluate the results of the RNA-seq, we selected seven DE-lncRNAs and seven DE-mRNAs related to lipid metabolism for qRT-PCR analyses. The qRT-PCR results were consistent with the RNA-seq data. (Table 3).

Table 3.
Comparison of qRT-PCR and RNA-seq results of 14 candidates (7 lncRNAs and 7 mRNAs).
3.7. Cis and Trans Roles of DE-lncRNAs
The potential functions of the 284 DE-lncRNAs were further explored by predicting their cis and trans targets. We found that 39 DE-lncRNAs were located adjacent to 65 protein-coding genes (Table S4), indicating that these lncRNAs might be associated with cis regulation of these genes. GO and KEGG enrichment analyses were conducted for these cis targets to better understand their functions and those of the cis-acting lncRNAs (Table S5). Regarding trans action, 3610 interactions were predicted to exist between 237 DE-lncRNAs and protein-coding genes (Table S4). Functional enrichment analysis found that these trans targets were significantly enriched in 147 GO terms and 20 KEGG pathways (Table S6), including several GO terms related to lipid metabolism, such as “lipid metabolic process”, “linoleic acid metabolic process”, and “unsaturated fatty acid biosynthetic process”. The pathways related to lipid metabolism included PPAR signaling, fatty acid degradation, and fatty acid metabolism pathways (Figure 5A). A heat map was constructed to highlight 10 selected target DE-mRNAs involved in these three pathways (Figure 5B), and an interaction network between these 10 lipid metabolism-related DE-mRNAs and DE-lncRNAs was generated using Cytoscape software (Figure 6). As shown in Figure 6, the trans-regulatory interaction network was quite complex. Some mRNAs (e.g., ehhadh encoding peroxisomal bifunctional enzyme, and slc25a48 encoding solute carrier family 25 member 48) were regulated by multiple lncRNAs (MSTRG.24713.1, MSTRG.7400.1, and others), and one lncRNA (MSTRG.30660.2) regulated multiple mRNAs (fabp7a encoding fatty acid-binding protein7a, faldh encoding fatty aldehyde dehydrogenase, and acsl3a encoding long chain fatty acid-CoA ligase 3a).
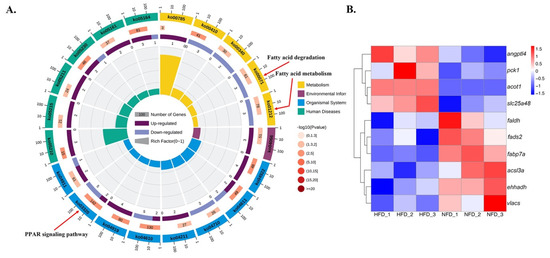
Figure 5.
(A) Results of KEGG enrichment analysis showing lipid metabolism-related pathways enriched with genes that are potentially trans-regulated by lncRNAs. (B) Heatmaps and hierarchical clustering of 10 selected mRNAs. Red, high expression; blue, low expression.
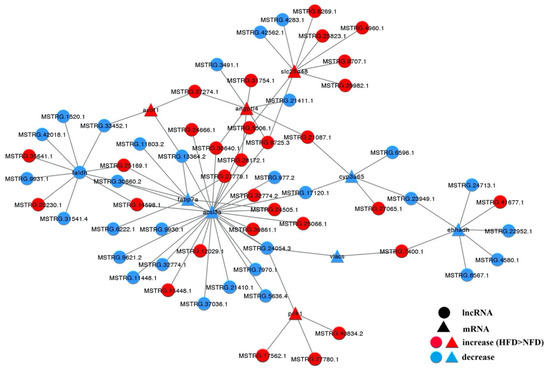
Figure 6.
LncRNA–mRNA interaction network in HFD vs. NFD groups. Ellipses represent lncRNAs, triangles represent mRNAs; red represents up-regulation (HFD vs. NFD) and blue represents down-regulation.
3.8. Expression Patterns of Several DE-lncRNAs and Their Lipid Metabolism-Related Target mRNAs in Response to a High-Fat Diet
On the basis of the results of the cis and trans action analyses, three lncRNAs (MSTRG.14598.1, MSTRG.6725.3, and MSTRG.13364.2) and their corresponding target mRNAs (faldh, slc25a48, and fabp7a) were selected for further analyses. Their expression patterns on days 14, 35, and 56 were determined (Figure 7). HFD exposure showed significantly up-regulated levels of MSTRG.14598.1 on day 14, day 35, and day 56 (p < 0.05) (Figure 7A), and similar trends in MSTRG.6725.3 (Figure 7B). The transcript level of MSTRG.13364.2 was not significantly different on days 14 and 35 (p > 0.05), but was significantly lower on day 56 after high-fat feeding (p < 0.05) (Figure 7C).
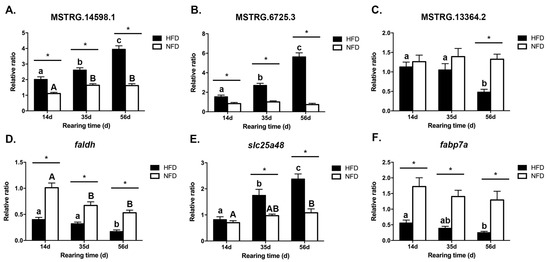
Figure 7.
Expression patterns of selected DE-lncRNAs and their lipid metabolism-related target mRNAs in the liver in the HFD group on days 14, 35, and 56. (n = 9). (A) MSTRG.14598.1; (B) MSTRG.6725.3; (C) MSTRG.13364.2; (D) faldh; (E) slc25a48; (F) fabp7a. * indicates significant difference (p < 0.05) between HFD and NFD groups at the same sampling time; different lowercase letters indicate significant difference (p < 0.05) in the HFD group among different sampling times. Different capital letters indicate significant difference (p < 0.05) in the NFD group among different sampling times.
4. Discussion
China is the world’s largest producer and trader of tilapia, although it is farmed worldwide [33]. Intensively farmed tilapia frequently suffer from fatty liver disease, which negatively affects their health and growth [4,5]. Previous studies have explored the regulatory mechanism of hepatic lipid metabolism in tilapia. For example, some key lipid metabolism genes have been functionally analyzed [34,35]. Our previous research suggested that miRNAs (e.g., miR-122, miR-29a, miR-145-5p) may be regulators of lipid metabolism in tilapia [4]. However, the functional roles of lncRNAs in tilapia were largely unknown. In this study, in an effort to better understand the roles of lncRNAs in lipid metabolism, we explored the hepatic lncRNA profiles of GIFT fed diets containing normal (8%) or high (18.5%) lipid levels using RNA-seq.
We used a rigorous filtering process to obtain 19,928 high-confidence lncRNAs from the livers of tilapia fed a HFD and an NFD. These lncRNAs had shorter transcript lengths and fewer exons compared with protein-coding mRNAs, consistent with the results of other studies on the lncRNAs of different freshwater fish [36,37]. We also identified 299 DE-mRNAs and 284 DE-lncRNAs in the liver between the HFD group and the NFD group. The reliability of RNA-seq data was confirmed by qRT-PCR. We found that the expression trend of all candidates was consistent with the RNA-seq results. However, there is a certain discrepancy between fold difference measured by qRT-PCR and RNA-seq analyses, and some reports found that these differences are normal and reasonable [38,39], which may be due to the different detection method between of sequencing and qRT-PCR and the relative low expression of candidate DE-lncRNAs in this study. Similar results have been reported previously [40,41], indicating that the identified candidates in the present study were reliable. Studies in non-fish animals have clearly demonstrated that some lncRNAs are critical regulators of lipid biosynthesis, lipid catabolism, and lipid transport to modulate lipid homeostasis [42,43]. Therefore, we speculate that these DE-lncRNAs identified here also affect liver lipid metabolism in tilapia.
LncRNAs can act as cis-regulators by targeting nearby protein-encoding genes [44]. Previous reports have documented some of the lncRNAs that play cis regulatory roles in hepatic lipid metabolism. For example, in chicken (Gallus domesticus), lncRNA-FNIP2 cis-regulates LPIN1, thereby regulating hepatic adipogenesis [45]. In blunt snout bream (Megalobrama amblycephala), lncRNA-MSTRG.6100 regulates two upstream cis targets, the genes encoding histone-lysine N-methyltransferase SET and MYND domain-containing protein 3, to regulate hepatic lipid metabolism [46]. Our analyses predicted 39 DE-lncRNAs with cis-acting regulation of 65 protein-coding genes. Identification of the functions of the targeted cis-regulated genes can provide clues about the regulatory functions of the DE-lncRNAs. LncRNAs can also regulate hepatic lipid metabolism via a trans-acting mechanism. For example, lncRNA-FLRL2 down-regulates the gene encoding the aryl hydrocarbon receptor nuclear translocator-like protein by a trans-acting mechanism, thereby participating in non-alcoholic fatty liver disease pathogenesis [47]. LncRNA-XLOC_001424 trans-regulates the expression of the gene encoding fatty acid desaturase 2 to promote non-alcoholic steatohepatitis in bama minipig (Sus scrofa domestica) [48]. In the present study, we identified 3610 pairs of DE-lncRNAs and trans-regulated targets. Thus, our data indicate that lncRNAs may participate in hepatic lipid metabolism in GIFT through cis- or trans-acting mechanisms.
Many pathways are related to lipid metabolism, such as the PPAR signaling pathway [49], the SREBP signaling pathway [50], the AMPK signaling pathway [51], and the fatty acid metabolism signaling pathway [52]. However, little was known about the roles of lncRNAs in regulating lipid metabolism in fish. The functions of lncRNAs are predicted mainly based on their target genes and related pathways [53]. In our analyses, we found that seven KEGG pathways were enriched with cis target genes and 20 KEGG pathways were enriched with trans target genes of the DE-lncRNAs. Several crucial pathways related to lipid metabolism were enriched with targets of DE-lncRNAs, including the PPAR signaling pathway, the fatty acid metabolism signaling pathway, the fatty acid degradation signaling pathway, and the alpha-linolenic acid metabolism signaling pathway. These findings strongly suggest that these are the key pathways by which DE-lncRNAs participate in the regulation of lipid metabolism.
To further investigate the specific roles of three lncRNAs (MSTRG.14598.1, MSTRG.6725.3 and MSTRG.13364.2) in lipid metabolism, we analyzed their expression patterns and those of their potential targets (faldh, slc25a48, and fabp7a), all of which are involved in key lipid metabolism signaling pathways, on day 14, day 35, and day 56. fabp7a and slc25a48 are two important regulators of the PPAR signaling pathway (Figure 8), and fabp7 is involved in fatty acid uptake and intracellular transport (Liu et al., 2021). Silencing of FABP7 was shown to relieve stearic acid-induced lipid accumulation in Hepa 16 cells [54]. slc25a48 encodes a member of the slc25 family that transports fatty acids across the mitochondrial membrane [55]. In this study, fabp7 was down-regulated and slc25a48 was up-regulated in the HFD group compared with the NFD group. Studies have shown that fish can change their lipid metabolism strategies, namely reduce lipogenesis and increase β-oxidation of fatty acids, to adapt to excessive lipid intake [56,57]. Therefore, in the HFD group, the down-regulation of fabp7 may have helped to reduce fatty acid uptake into hepatocytes, while the up-regulation of slc25a48 may have enhanced fatty acid transport to the mitochondria for β-oxidation. Interestingly, we detected positive relationships between the levels of the lncRNAs MSTRG.6725.3 or MSTRG.13364.2 and those of slc25a48 and fabp7a transcripts, respectively, in the HFD group. LncRNAs bind to the 3′-untranslated (UTR) region of the target gene’s mRNA and block a large number of miRNA target sites, thereby maintaining the stability of the target mRNA [58]. LncRNA-PXN-AS1-L was suggested to bind to the 3-′UTR of PXN mRNA, thereby stabilizing it and increasing PXN expression, ultimately promoting the growth of tumor cells [59]. We speculate that MSTRG.6725.3 and MSTRG.13364.2 may stabilize transcripts of their target genes slc25a48 and fabp7a, respectively, thereby positively regulating their expression and maintaining lipid homeostasis.
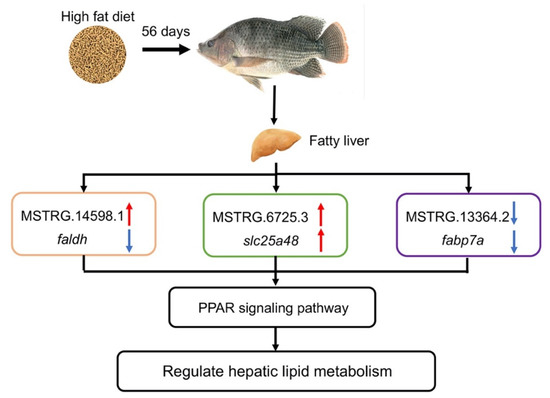
Figure 8.
Diagram of lncRNA-mediated regulated pathway in the liver of GIFT fed a HFD for 56 days.
FALDH is a crucial enzyme for phytic acid decomposition [60]. Activation of FALDH can protect cells from oxidative stress associated with lipid peroxidation [60]. The expression of FALDH is regulated by the PPARα signaling pathway [60]. In the present study, faldh was down-regulated in the HFD group, which had disrupted hepatic lipid metabolism. This may have led to the accumulation of lipid peroxides and, consequently, oxidative stress injury. LncRNAs can form complementary double strands with transcripts of protein-encoding genes, and produce endogenous siRNAs under the action of Dicer enzymes. In this way, they can silence gene expression [58]. In this study, MSTRG.14598.1 was up-regulated and faldh was down-regulated in the HFD group, indicating that the interaction between MSTRG.14598.1 and faldh might play a crucial role in hepatic fatty acid metabolism.
5. Conclusions
To our knowledge, this is the first report on the differences in hepatic lncRNA-mRNA expression profiles between GIFT fed a NFD and those fed a HFD. A total of 299 DE-mRNAs and 284 DE-lncRNAs were identified based on RNA-seq data analyses. The transcript profiles of seven DE-lncRNAs and seven DE-mRNAs related to lipid metabolism were verified by qRT-PCR. Functional enrichment analyses of cis- and trans-targets of DE-lncRNAs revealed that these DE-lncRNAs were involved in regulating key signaling pathways in lipid metabolism, such as PPAR signaling, fatty acid degradation, and fatty acid metabolism. Analyses of the expression patterns of three lncRNAs (MSTRG.14598.1, MSTRG.6725.3, and MSTRG.13364.2) and their potential targets (faldh, slc25a48, and fabp7a) revealed that these lncRNAs might participate in lipid metabolism by post-transcriptionally regulating their respective target genes. Although further experimental analyses are required to test the interactions between these lncRNAs and mRNAs, the information gained in this study provides new insights into the molecular mechanism of lncRNA-mediated lipid metabolism in tilapia. The lncRNAs identified in this study will serve as a reference for further research on the regulation of lipid metabolism in tilapia.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes7060332/s1, Figure S1: Density plot (A) showing the distribution of expression density of each sample. Box plot (B) showing the distribution of maximum, minimun and percentile values for normalized signals of each sample; Figure S2: DE-mRNAs enriched in three categories: biological process, molecular function and cellular component, and their main level 2 GO terms. Table S1: Ingredients and composition of NFD and HFD; Table S2: Overview of hepatic sequencing data and quality filtering.
Author Contributions
P.X. and J.Q. conceived and designed the experiments, S.L. performed the experiments. T.Z. analyzed the RNA-seq data. M.L. conducted qRT-PCR experiments. Y.T. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Special Fund of the National Natural Science Foundation of China (32002363) and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (2020TD37).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Bioethical Committee of the Freshwater Fisheries Research Center (FFRC), Chinese Academy of Fishery Sciences (2013863BCE).
Data Availability Statement
All raw sequencing data have been deposited in NCBI and Sequence Read Archive databases under the accession number GSE 211712 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE211712) (accessed on 21 August 2022) with secure token “gxyrougctbofhcj”.
Acknowledgments
We thank Jennifer Smith for editing the English text of various drafts of this manuscript.
Conflicts of Interest
The authors declare that they have no competing interest.
References
- Xu, H.; Ren, M.C.; Liang, H.L.; Ge, X.P.; Ji, K.; Huang, D.Y.; Yu, H.; Wu, L.H. Interactive effects of water salinity and dietary methionine levels on growth performance, whole-body composition, plasma parameters, and expression of major nutrient metabolism genes in juvenile genetically improved farmed Tilapia (Oreochromis niloticus). Aquaculture 2022, 546, 737381. [Google Scholar] [CrossRef]
- Qiang, J.; Tao, Y.F.; Bao, J.W.; Chen, D.J.; Li, H.X.; He, J.; Xu, P. High Fat Diet-Induced miR-122 Regulates Lipid Metabolism and Fat Deposition in Genetically Improved Farmed Tilapia (GIFT, Oreochromis niloticus) Liver. Front. Physiol. 2018, 9, 1422. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.Y. Causes of fatty liver in farmed fish: A review and new perspectives. J. Fish. China 2014, 38, 1628–1638. [Google Scholar]
- Tao, Y.F.; Qiang, J.; Yin, G.J.; Xu, P.; Shi, Q.; Bao, J.W. Identification and characterization of lipid metabolism-related microRNAs in the liver of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) by deep sequencing. Fish. Shellfish Immun. 2017, 69, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Cao, L.P.; Du, J.L.; He, Q.; Gu, Z.Y.; Jeney, G.; Xu, P.; Yin, G.J. Effects of High-Fat Diet on Steatosis, Endoplasmic Reticulum Stress and Autophagy in Liver of Tilapia (Oreochromis niloticus). Front. Mar. Sci. 2020, 7, 363. [Google Scholar] [CrossRef]
- Lu, K.L.; Xu, W.N.; Li, J.Y.; Li, X.F.; Huang, G.Q.; Liu, W.B. Alterations of liver histology and blood biochemistry in blunt snout bream Megalobrama amblycephala fed high-fat diets. Fish. Sci. 2013, 79, 661–671. [Google Scholar] [CrossRef]
- Qiang, J.; He, J.; Yang, H.; Sun, Y.L.; Tao, Y.F.; Xu, P.; Zhu, Z.X. Dietary lipid requirements of larval genetically improved farmed tilapia, Oreochromis niloticus (L.), and effects on growth performance, expression of digestive enzyme genes, and immune response. Aquac. Res. 2017, 48, 2827–2840. [Google Scholar] [CrossRef]
- Qiang, J.; Khamis, O.A.M.; Jiang, H.J.; Cao, Z.M.; He, J.; Tao, Y.F.; Xu, P.; Bao, J.W. Effects of dietary supplementation with apple peel powder on the growth, blood and liver parameters, and transcriptome of genetically improved farmed tilapia (GIFT, Oreochromis niloticus). PLoS ONE 2019, 14, e0224995. [Google Scholar] [CrossRef]
- Tian, J.; Wen, H.; Zeng, L.B.; Jiang, M.; Wu, F.; Liu, W.; Yang, C.G. Changes in the activities and mRNA expression levels of lipoprotein lipase (LPL), hormone-sensitive lipase (HSL) and fatty acid synthetase (FAS) of Nile tilapia (Oreochromis niloticus) during fasting and re-feeding. Aquaculture 2013, 400, 29–35. [Google Scholar] [CrossRef]
- He, A.Y.; Ning, L.J.; Chen, L.Q.; Chen, Y.L.; Xing, Q.; Li, J.M.; Qiao, F.; Li, D.L.; Zhang, M.L.; Du, Z.Y. Systemic adaptation of lipid metabolism in response to low- and high-fat diet in Nile tilapia (Oreochromis niloticus). Physiol. Rep. 2015, 3, e12485. [Google Scholar] [CrossRef]
- Ma, X.Y.; Qiang, J.; He, J.; Gabriel, N.N.; Xu, P. Changes in the physiological parameters, fatty acid metabolism, and SCD activity and expression in juvenile GIFT tilapia (Oreochromis niloticus) reared at three different temperatures. Fish Physiol. Biochem. 2015, 41, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Moran, V.A.; Perera, R.J.; Khalil, A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012, 40, 6391–6400. [Google Scholar] [CrossRef]
- Kapusta, A.; Feschotte, C. Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends. Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef]
- Wang, X. Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J. Cell. Biochem. 2018, 119, 1567–1574. [Google Scholar] [CrossRef]
- Li, D.; Guo, L.; Deng, B.; Li, M.; Yang, T.; Yang, F.; Yang, Z. Long non-coding RNA HR1 participates in the expression of SREBP-1c through phosphorylation of the PDK1/AKT/FoxO1 pathway. Mol. Med. Rep. 2018, 18, 2850–2856. [Google Scholar] [CrossRef]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A Liver-Enriched Long Non-Coding RNA, lncLSTR, Regulates Systemic Lipid Metabolism in Mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef]
- Xu, H.G.; Cao, L.; Sun, B.; Wei, Y.L.; Liang, M.Q. Transcriptomic Analysis of Potential “IncRNA-mRNA” Interactions in Liver of the Marine Teleost Cynoglossus semilaevis Fed Diets With Different DHA/EPA Ratios. Front. Physiol. 2019, 10, 331. [Google Scholar] [CrossRef]
- Sehgal, P.; Mathew, S.; Sivadas, A.; Ray, A.; Tanwar, J.; Vishwakarma, S.; Ranjan, G.; Shamsudheen, K.V.; Bhoyar, R.C.; Pateria, A.; et al. LncRNA VEAL2 regulates PRKCB2 to modulate endothelial permeability in diabetic retinopathy. EMBO J. 2021, 40, e107134. [Google Scholar] [CrossRef]
- Tao, Y.F.; Qiang, J.; He, J.; Zhu, H.J.; Bao, J.W.; Xu, P. Untargeted LC-MS metabolomics approach reveals metabolic changes in genetically improved farmed tilapia (Oreochromis niloticus) with fatty liver induced by a high-fat diet. Aquac. Res. 2021, 52, 724–735. [Google Scholar] [CrossRef]
- Li, M.X.; Qiang, J.; Zhu, X.W.; Bao, J.W.; Tao, Y.F.; Zhu, H.J. Effect of Siberian Ginseng Water Extract as a Dietary Additive on Growth Performance, Blood Biochemical Indexes, Lipid Metabolism, and Expression of PPARs Pathway-Related Genes in Genetically Improved Farmed Tilapia (Oreochromis niloticus). Fishes 2022, 7, 149. [Google Scholar] [CrossRef]
- Wang, L.M.; Zhu, W.B.; Dong, Z.J.; Song, F.B.; Dong, J.J.; Fu, J.J. Comparative microRNA-seq Analysis Depicts Candidate miRNAs Involved in Skin Color Differentiation in Red Tilapia. Int. J. Mol. Sci. 2018, 19, 1209. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.T.; Bu, D.C.; Zhao, G.G.; Yu, K.; Zhang, C.H.; Liu, Y.N.; Chen, R.S.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- He, F.; Liu, Q.Q.; Zheng, L.; Cui, Y.Q.; Zheng, L.Q. RNA-Seq Analysis of Rice Roots Reveals the Involvement of Post-Transcriptional Regulation in Response to Cadmium Stress. Front. Plant Sci. 2016, 6, 1136. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Quan, J.Q.; Kang, Y.J.; Luo, Z.C.; Zhao, G.Y.; Ma, F.; Li, L.L.; Liu, Z. Identification and characterization of long noncoding RNAs provide insight into the regulation of gene expression in response to heat stress in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. D 2020, 36, 100707. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Alkan, F.; Wenzel, A.; Palasca, O.; Kerpedjiev, P.; Rudebeck, A.F.; Stadler, P.F.; Hofacker, I.L.; Gorodkin, J. RIsearch2: Suffix array-based large-scale prediction of RNA-RNA interactions and siRNA off-targets. Nucleic Acids Res. 2017, 45, e60. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Jiang, W.; Jiang, Y.; Wang, S.; Fang, J.; Zhu, L.; Zhu, Y.; Yan, G.; Sun, H.; Chen, L.; et al. Preliminary functional inquiry of lncRNA ENST00000433673 in embryo implantation using bioinformatics analysis. Syst. Biol. Reprod. Med. 2019, 65, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Abbas, G.; Ghaffar, A.; Ferrando, S.; Gallus, L. Effect of Different Salinity Level on Breeding, Fertilization, Hatching and Survival of Nile Tilapia, Oreochromis niloticus (Linnaeus, 1758) in Captivity. Pak. J. Zool. 2018, 50, 539–547. [Google Scholar] [CrossRef]
- He, A.Y.; Liu, C.Z.; Chen, L.Q.; Ning, L.J.; Qin, J.G.; Li, J.M.; Zhang, M.L.; Du, Z.Y. Molecular characterization, transcriptional activity and nutritional regulation of peroxisome proliferator activated receptor gamma in Nile tilapia (Oreochromis niloticus). Gen. Comp. Endocrinol. 2015, 223, 139–147. [Google Scholar] [CrossRef]
- Ning, L.J.; He, A.Y.; Li, J.M.; Lu, D.L.; Jiao, J.G.; Li, L.Y.; Li, D.L.; Zhang, M.L.; Chen, L.Q.; Du, Z.Y.; et al. Mechanisms and metabolic regulation of PPARα activation in Nile tilapia (Oreochromis niloticus). BBA-Mol. Cell Biol. L. 2016, 1861, 1036–1048. [Google Scholar] [CrossRef]
- Luo, M.K.; Wang, L.M.; Yin, H.R.; Zhu, W.B.; Dong, Z.J. Integrated analysis of long non-coding RNA and mRNA expression in different colored skin of koi carp. BMC Genom. 2019, 20, 515. [Google Scholar] [CrossRef]
- Gan, L.; Wang, Y.Z.; Chen, S.J.; Lin, Z.H.; Sun, J.J.; He, Y.H.; Tang, H.J.; Peng, J.; Guo, H.H. Identification and characterization of long non-coding RNAs in muscle sclerosis of grass carp, Ctenopharyngodon idellus fed with faba bean meal. Aquaculture 2020, 516, 734521. [Google Scholar] [CrossRef]
- Su, Z.; Labaj, P.P.; Li, S.; Thierry Mieg, J.; Thierry Mieg, D.; Shi, W.; Wang, C.; Schroth, G.P.; Setterquist, R.A.; Thompson, J.F.; et al. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014, 32, 903–914. [Google Scholar]
- Zhu, W.; Wang, L.; Dong, Z.; Chen, X.; Song, F.; Liu, N.; Yang, H.; Fu, J. Comparative Transcriptome Analysis Identifies Candidate Genes Related to Skin Color Differentiation in Red Tilapia. Sci. Rep. 2016, 6, 31347. [Google Scholar] [CrossRef]
- Li, H.; Cui, P.; Fu, X.; Zhang, L.; Yan, W.; Zhai, Y.; Lei, C.; Wang, H.; Yang, X. Identification and analysis of long non-coding RNAs and mRNAs in chicken macrophages infected with avian infectious bronchitis coronavirus. BMC Genomics 2021, 22, 67. [Google Scholar] [CrossRef]
- Sun, X.; Jia, B.; Qiu, X.L.; Chu, H.X.; Zhang, Z.Q.; Wang, Z.P.; Zhao, J.J. Potential functions of long non-coding RNAs in the osteogenic differentiation of human bone marrow mesenchymal stem cells. Mol. Med. Rep. 2019, 19, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.N.; Wang, W.; Yang, N.; Huang, X.M.; Liu, C.F. Regulation of Glucose and Lipid Metabolism by Long Non-coding RNAs: Facts and Research Progress. Front. Endocrinol. 2020, 11, 457. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Hwang, H.J.; Cho, J.Y. Long Non-Coding RNA Associated with Cholesterol Homeostasis and Its Involvement in Metabolic Diseases. Int. J. Mol. Sci. 2020, 21, 8337. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.; Vincendeau, M.; Eitelhuber, A.C.; Krappmann, D. Mechanisms and consequences of constitutive NF-kappa B activation in B-cell lymphoid malignancies. Oncogene 2014, 33, 5655–5665. [Google Scholar] [CrossRef]
- Guo, L.J.; Chao, X.H.; Huang, W.L.; Li, Z.H.; Luan, K.; Ye, M.; Zhang, S.Y.; Liu, M.Q.; Li, H.M.; Luo, W.; et al. Whole Transcriptome Analysis Reveals a Potential Regulatory Mechanism of LncRNA-FNIP2/miR-24-3p/FNIP2 Axis in Chicken Adipogenesis. Front. Cell Dev. Biol. 2021, 9, 653798. [Google Scholar] [CrossRef]
- Jia, X.Y.; He, C.; Jiang, W.B.; Wen, C.; Gao, F.; Jiang, G.Z.; Li, X.F.; Chi, C.; Liu, W.B.; Zhang, D.D. Identification of potential pathways whereby dietary L-tryptophan ameliorates the glucose metabolic disorder of blunt snout bream through long non-coding RNAs. Aquaculture 2021, 545, 737256. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, H.-x.; Xu, C.-f.; Yu, C.-h.; Li, Y.-m. Long Non-Coding RNA Profiling in a Non-Alcoholic Fatty Liver Disease Rodent Model: New Insight into Pathogenesis. Int. J. Mol. Sci. 2017, 18, 21. [Google Scholar] [CrossRef]
- Xia, J.H.; Xin, L.L.; Zhu, W.J.; Li, L.; Li, C.X.; Wang, Y.F.; Mu, Y.L.; Yang, S.L.; Li, K. Characterization of long non-coding RNA transcriptome in high-energy diet induced nonalcoholic steatohepatitis minipigs. Sci. Rep. 2016, 6, 30709. [Google Scholar] [CrossRef]
- Hu, N.; Chen, C.Y.; Wang, J.H.; Huang, J.; Yao, D.H.; Li, C.L. Atorvastatin Ester Regulates Lipid Metabolism in Hyperlipidemia Rats via the PPAR-signaling Pathway and HMGCR Expression in the Liver. Int. J. Mol. Sci. 2021, 22, 11107. [Google Scholar] [CrossRef]
- Ma, Y.J.; Xu, L.Y.; Rodriguez-Agudo, D.; Li, X.B.; Heuman, D.M.; Hylemon, P.B.; Pandak, W.M.; Ren, S.L. 25-Hydroxycholesterol-3-sulfate regulates macrophage lipid metabolism via the LXR/SREBP-1 signaling pathway. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E1369–E1379. [Google Scholar] [CrossRef]
- Li, Y.; Ding, H.; Dong, J.; Rahman, S.U.; Feng, S.; Wang, X.; Wu, J.; Wang, Z.; Liu, G.; Li, X.; et al. Glucagon attenuates lipid accumulation in cow hepatocytes through AMPK signaling pathway activation. J. Cell. Physiol. 2019, 234, 6054–6066. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.G.; Pang, D.X.; Lu, C.; Xu, A.; Huang, P.X.; Ouyang, H.S.; Yu, H. Investigation on the Effect of Two Fat Metabolism Related Pathways on Intramuscular Fat Content in Pigs. Pak. J. Zool. 2021, 53, 1353–1366. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Lee, H.; Jung, C.H.; Ha, T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol. Nutr. Food Res. 2012, 56, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Martyniuk, C.J.; Buerger, A.N.; Vespalcova, H.; Rudzanova, B.; Sohag, S.R.; Hanlon, A.T.; Ginn, P.E.; Craft, S.L.; Smetanova, S.; Budinska, E.; et al. Sex-dependent host-microbiome dynamics in zebrafish: Implications for toxicology and gastrointestinal physiology. Comp. Biochem. Physiol. D. 2022, 42, 100993. [Google Scholar] [CrossRef]
- Li, A.X.; Yuan, X.C.; Liang, X.F.; Liu, L.W.; Li, J.; Li, B.; Fang, J.G.; Li, J.; He, S.; Xue, M.; et al. Adaptations of lipid metabolism and food intake in response to low and high fat diets in juvenile grass carp (Ctenopharyngodon idellus). Aquaculture 2016, 457, 43–49. [Google Scholar] [CrossRef]
- Leng, X.J.; Wu, X.F.; Tian, J.; Li, X.Q.; Guan, L.; Weng, D.C. Molecular cloning of fatty acid synthase from grass carp (Ctenopharyngodon idella) and the regulation of its expression by dietary fat level. Aquacult. Nutr. 2012, 18, 551–558. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Yuan, J.H.; Liu, X.N.; Wang, T.T.; Pan, W.; Tao, Q.F.; Zhou, W.P.; Wang, F.; Sun, S.H. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 2017, 19, 820–832. [Google Scholar] [CrossRef]
- Verhoeven, N.M.; Jakobs, C.; Carney, G.; Somers, M.P.; Wanders, R.J.A.; Rizzo, W.B. Involvement of microsomal fatty aldehyde dehydrogenase in the alpha-oxidation of phytanic acid. FEBS Lett. 1998, 429, 225–228. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).